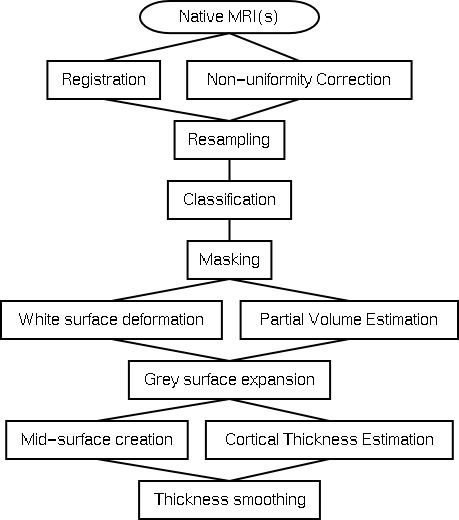
The Image Processing Pipeline
The image processing pipeline incorporates all of the necessary steps to go from one or more MRIs from the scanner all the way to output data ready for statistical analysis. At this point in time the key output is a smoothed cortical thickness map. The Statistics page contains the information about how to process thickness data for population studies.
Below is the data processing tree in a clickable map:
Native Files
Each subject needs at least one MRI, though more are possible. The most common case is a single T1 weighted scan, often a T2 and PD scan are included as well. The key components are the resolution, signal to noise, and ability to classify the cortical grey matter and extra cortical cerebro-spinal fluid. Since the cortex is usually between one and four millimetres resolution is especially important.The files need also be convertable to the MINC format.
Registration
The registration step occurs at the same time as the non-uniformity correction step. At this point only a nine parameter linear registration is performed to align the subject into standardised stereotaxic space. If the subject has more than one MRI, the different modalities are registered to each other using mutual information criterion and then aligned into stereotaxic space.
Non-uniformity Correction
Non-uniformity correction, performed using the N3 algorithm, is designed to remove inhomogeneity fields introduced by the scanner. This step is repeated separately for each scan of each subject.
Resampling
Here the transform to stereotaxic space is used to resample the non-uniformity corrected file.
Classification
The non-uniformity corrected files in stereotaxic space are classified into four tissues: grey matter, white matter, cerebro-spinal fluid, and background. One or more image modalities can be used to achieve this classification. The INSECT algorithm is used.
Masking
The skull, brainstem, cerebellum, and dura are removed through the deformation of a fairly rigid sphere onto the classified volume.
White surface deformation
An ellipsoid is deformed in an iterative fashion onto the classified white matter. This is a computationally very complex step that results in a white matter surface of spherical topology consisting of 81920 vertices. This step is part of the ASP algorithm.
Partial Volume Estimation
Each voxel is modeled as containing some proportion of the three main tissue classes - white matter, grey matter, and cerebro-spinal fluid. This technique can use either a T1 or a T1 plus a T2 and PD.
Grey surface expansion
Once the white matter surface has been extracted, it is used as a starting point for expansion out to the grey surface. Topological correctness is maintained throughout this deformation process. This is also part of the ASP algorithm.
Mid surface creation
For visualisation and blurring purposes a mid-cortical surface is created through geometric averaging of the white and grey surfaces.
Cortical Thickness Estimation
Once the three surfaces exist cortical thickness can be estimated at every vertex using - at this point - one of six different algorithms.
Thickness smoothing
Once cortical thickness is computed the values are smoothed using a contour following blurring algorithm developed by Moo Chung.