Try to download these small minc volumes and object files to test the Display software. Display
is a 3D viusalisation sofware developped by David
MacDonald .
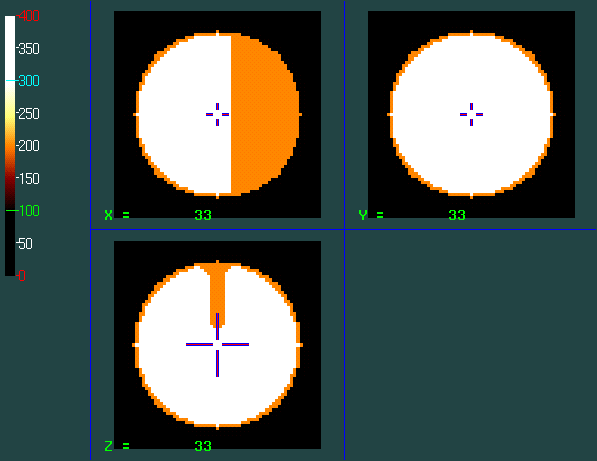
A simple minc volume :

This is a 67*67*67 3D volume. Download it final_t1_MY.mnc.gz (7 KB, compressed with gzip)
A simple geometrical object (a sphere representing the external surface of the 3D volume) :

This surface is composed of 12776 polygons. Download surface_MY.obj.gz (164 K, compressed with gzip)
A 3D T1 MRI (raw data format):
file: final_t1_02.raw.gz
image: signed_short 0 to 4095
image dimensions: zspace yspace xspace
| dimension name | length | step | start |
| zspace | 181 | 1 | -72 |
| yspace | 217 | 1 | -126 |
| xspace | 181 | 1 | -90 |
z increases from inferior to superior
y increases from posterior to anaterior
x increases from left to right
Cortical Surface .pl (under construction)
Tar file:CSPL.tar.gz (IRIX 6.5)
Once the file is unzipped (gunzip) and untarred (tar -xvf), edit the cortical_surface.pl file to set the correct paths (look for string '#change path here:' in the file).
Send an e-mail georges@bic.mni.mcgill.ca if you dowload this program
Usage: cortical_surface.pl volume.mnc output_file.obj threshold_value [transform.xfm]
Option: If the volume is not in Talairach space, a transform must be specified ([transform.xfm])
Quick help: cortical_surface.pl deforms the model cortical surface (stored in ./Deform) such that this surface fits the Grey Matter (GM)/CSF interface. Then threshold_valued should be equal to a minc real value (Vl) in between the GM mean real value and CSF mean real value. This value may change from a MRI volume to another. The program works only if the 3D MRI is resampled into the Talairach space (linear registration is performed using mritotal)
This program was created by David MacDonald
(Note that there is another version with conglomerate's link).
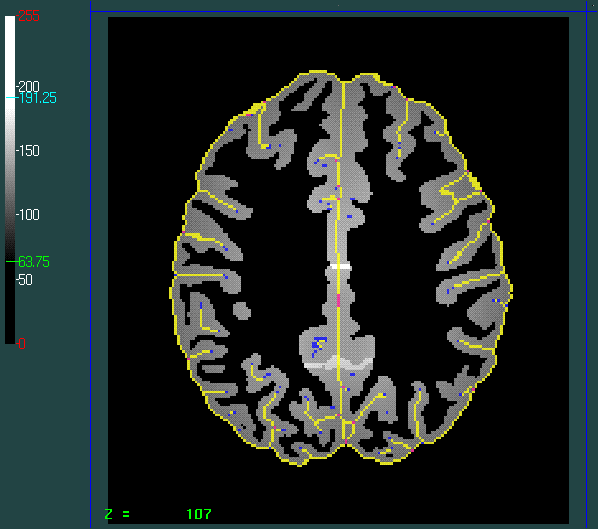
Skeleton Test
Grey Matter : GM; White Matter: WM; CerebroSpinal Fluid: CSF
Region of Interest (ROI) = ((GM+WM)*C)\WM
where *C : is a morphological closing; \ : is the difference between sets
Value of the external boundary of the ROI: 113
Slice : Zworld =35; Zvoxel = 107
Superimposed is the skeleton computed by removing simple points. The skeleton colors correspond to topological labels: yellow: surface; blue: simple point; pink: surface junctions. This skeleton is not optimal for our application because it is sensitive to noise and because important sulci may be forgotten in the final skeleton.
ROI homotopic: roi_homotopic.mnc.gz ; Skeleton: skel_homotopic.mnc.gz
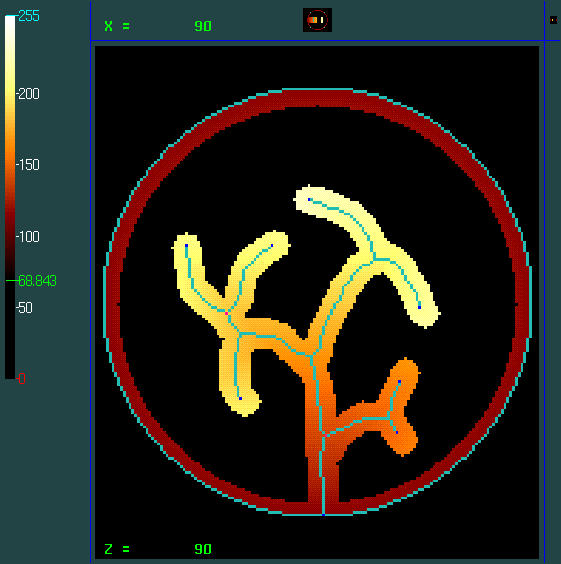
Value of the external boundary of the ROI: 113
Slice: Zworld = 18; Zvoxel = 90.
Simulated ROI : roi_homotopic_simulated.mnc.gz ;
Simulated skeleton: skel_homotopic_simulated.mnc.gz ; light blue: surface; blue: simple point; pink: surface junctions.
DON'T FORGET THAT THESE FILES ARE ZIPPED WITH GZIP
(don't forget the .gz extension)
If you have comments or suggestions, email me at georges@bic.mni.mcgill.ca