mincFiberTrack Man page
NAME
mincFiberTrack - does fiber tracking using various algorithms
SYNOPSIS
mincFiberTrack [<options>]
DESCRIPTION
mincFiberTrack implements various tracking algorithms, namely the most commonly used FACT(Fiber Assignment by Continuous Tracking) algorithm as well as others which use high angular resolution data to reconstruct more than one direction per voxel. The function uses the output from minctensor. The tracking can be limited by a fractional anisotropy threshold and/or a curvature constraint, as well as through inclusion/exclusion masks. The outputs are a binary connectivity map and/or a VTK file used to visualize the tracts with minc3Dvis.
OPTIONS
Usage: FibreTrack [options]
algorithm options:
-algorithm: FACT, Maxima, Probabilistic. The following summarizes these algorithm choices:
FACT: tracking follows the principal eigenvector of the diffusion tensor - no quantification of uncertainty
Maxima: tracking follows one of multiple fibre directions that have been obtained using a high angular resolution approach. The fibre direction closest to the incoming fibre direction is selected.
Probabilistic: the tracking incorporates the output of ProbDeconvolve, which labels voxels as containing single fibres or double or triple
crossings. This is done in a probabilistic framework. (Notes: (1) This option is non-probabilistic if the input file has no uncertainty in
it. (2) The code supports the output of a program called ProbLabels, which adds fanning to the possible voxel geometries, but this program
is not yet part of the mincdiffusion tools.)
FACT:
-e1x <e1x.mnc> -e1y <e1y.mnc> -e1z <e1z.mnc>
or: -namebase <namebase>
Maxima:
-ODF <ODF_filename.mnc>
-ODF_maxima <maxima_filename>: optionally input these if already calculated: can only input them if use -noresample option
Probabilistic:
-labels <labels.mnc>: the file must be of the format output by mincProbDeconvolve: see mincProbDeconvolve -help for details.
output files and options:
-tracts <output_tracts.vtk>. This option is highly discouraged for the Probabilistic algorithm, and for the FACT and Maxima algorithms if doing extensive brute force tractography (see below for the brute_force option).
-conn <binary_connectivity_map.mnc>: 1 where tracking went, 0 where it didn't go
-conn_index <connectivity_index_filename.mnc>: (for Probabilistic algorithm): at each voxel tract passes through, write the connectivity index for the tract segment between the reference ROI (see above) and that voxel.
-binary: save vtk file in binery (default ascii)
-obj: output tracts in MINC .obj format rather than vtk format
-nobbox: save full-size output minc files [default: smallest bounding box]
-clobber: overwrite existing output file
other options for all algorithms:
-brute_force: start at all voxels and check whether went through ROIs
-mask <mask.mnc> -(gt,lt,ge,le) <threshold> (only one comparison allowed per mask)
-roi <roi.mnc> (all values greater than 0 are treated is being inside the roi). multiple -roi inputs may be given; tracts that pass through ALL ROIs are retained
-reference_roi <integer>: integer corresponding to the -roi in the input list that connectivity index values (cumulative minimum ODF tangent to tracts; see below) are relative to. default: 1
-exclude <exclusion_mask.mnc>: remove all tracts that pass through this ROI
-curvature_constraint_angle <angle in degrees, applied for each step regardless of -step input
-roi_t <roi.txt> -(v,w) : text file listing coordinates of roi (x1 y1 z1\n ... xn yn z\n) in either voxel(v) or world (w) coordinates
-max_length <length>: maximum tract length in mm
-step <step>: grid size for tracking. default is the voxel sampling, and this is generally recommended for speed when using brute_force option
-tmpdir: a temporary directory name (to be created by the program). Default is /tmp/tmpXXXXXXXXX
-save_temp_dir: save the temporary directory and resampled files (so can run without resampling in the future if desired)
-noresample: don't resample any input files: assume in same space
-iterations: iterations of algorithm (default: 10000) (Probabilistic algorithm)
-seedfreq <freq>: number of starts per voxel per dimension (default: 3)
Generic options
-help: Print summary of command-line options and abort.
TIPS
For the fibertracking results to make sense, it is useful to use termination criteria. FA threshold -> only tract in voxels where FA is above a threshold i.e. significant directionality curvature constraint angle -> stop tracking if curvature from one voxel to the next is above a threshold (e.g. 70 degrees), this is to help avoid jumping to another tract brute force: this starts tracking in all voxels and only keeps those passing through the roi. The figure below shows an example of how tracking results can differ between the brute force approach and only seeding in the roi.
The lines represent the principal eigenvector direction in each voxel. More branches of the tract are picked up with the brute force approach (green).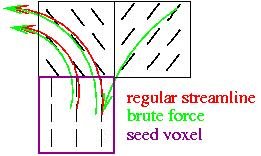
EXAMPLES
1- Start FACT fibertracking from a seed region (
ROI.mnc) and continue tracking only if the FA value is above 0.2 and the curvature of the tract is less than 70 degrees. The input files (infile-reg-with-t1-mask_e1x.mnc,infile-reg-with-t1-mask_e1y.mnc,infile-reg-with-t1-mask_e1z.mnc,infile-reg-with-t1-mask_FA.mnc) are the outputs from the preprocessing step tensor_preprocess followed by tensor calculation with minctensor.First draw a seed region ROI using
Display, then:
mincFibreTrack -algorithm FACT -namebase infile-reg-with-t1-mask -mask infile-reg-with-t1-mask_FA.mnc -gt 0.2 -roi ROI.mnc -curvature_constraint_angle 70 -conn conn_map.mnc -tracts output_tracts.vtk2- Do an exhaustive search (i.e. seed in every voxel of the volume) to find which tracts pass through
ROI1.mncBUT NOT throughROI2.mnc(same stopping criteria as above for FA and curvature):mincFibreTrack -algorithm FACT -brute_force -namebase infile-reg-with-t1-mask -mask infile-reg-with-t1-mask_FA.mnc -gt 0.2 -roi ROI1.mnc -exclude ROI2.mnc -curvature_constraint_angle 70 -conn conn_map.mnc -tracts output_tracts.vtk3- Do an exhaustive search (i.e. seed in every voxel of the volume) to find which tracts pass through
ROI1.mncAND throughROI2.mnc(same stopping criteria as above for FA and curvature):NOTE: IfmincFibreTrack -algorithm FACT -brute_force -namebase infile-reg-with-t1-mask -mask infile-reg-with-t1-mask_FA.mnc -gt 0.2 -roi ROI1.mnc -roi ROI2.mnc -curvature_constraint_angle 70 -conn conn_map.mnc -tracts output_tracts.vtkROI1.mncandROI2.mncwere kept in the same file, all voxel in each roi would be used as a seed, this would be like keeping tracts that pass throughROI1.mncORROI2.mnc4- Track using multiple fibers per voxel (using mincDeconvolve), initiating in a text roi specified in world coordinates (same stopping criteria as above for FA and curvature):
NOTE:test-roi.txtlooks something like this:
-11.9639 54.8125 80.066
-11.665 44.511 59.9207
-5.71236 5.83727 58.9366
-1.57408 29.1042 42.2982
-17.2905 -17.2855 44.1643
-21.6102 -35.9768 20.4383
mincFibreTrack -algorithm Maxima -ODF deconvolve.mnc -mask infile-reg-with-t1-mask_FA.mnc -gt 0.2 -roi_t text-roi.txt -w -curvature_constraint_angle 70 -conn conn_map.mnc -tracts output_tracts.vtk5- Track using multiple fibers per voxel and get the strength of connection to the seed ROI (using mincProbDeconvolve).
mincFibreTrack -algorithm Probabilistic -labels probdeconvolve.mnc -mask infile-reg-with-t1_FA.mnc -gt 0.1 -mask infile-reg-with-t1_MD.mnc\ -lt 1.0 -roi roi.mnc -curvature_constraint_angle 80 -iterations 5000 -conn_index conn_index_probdeconv.mnc
NOTE: Don't use the-tractsoption, it will run FOREVER!
Ilana LEPPERT
created: 21st March 2007
last modified: 24th March 2011