DL Collins, AP Zijdenbos and AC Evans
McConnell Brain Imaging Centre, Montréal Neurological
Institute,
McGill University, Montréal, Canada
Poster presented at the 4-th International Conference on Functional Mapping of the Human Brain
DL Collins, AP Zijdenbos and AC Evans
McConnell Brain Imaging Centre, Montréal Neurological
Institute,
McGill University, Montréal, Canada
Quantitative analysis of neuro-anatomical or neuro-functional data often requires explicit regional identification of gross anatomical structures. Unfortunately, manual segmentation is time-consuming, subjective and error prone. Furthermore, inter- and intra-observer variability may reduce detectability of subtle differences when making comparisons. To address this problem, we have developed two complementary segmentation strategies for automatic identification of tissue types and gross anatomical structures from volumetric magnetic resonance images (MRI) of the human brain.
The first, named ![]() (Automatic Nonlinear Image Matching and Anatomical
Labeling, [1]), was designed to segment anatomical structures
by computing a non-linear warp to register a given subject's MRI to a
pre-labeled target MRI that serves as an atlas. The inverse of the
recovered transformation is used to warp the atlas labels back onto the
subject's MRI, thus achieving segmentation.
(Automatic Nonlinear Image Matching and Anatomical
Labeling, [1]), was designed to segment anatomical structures
by computing a non-linear warp to register a given subject's MRI to a
pre-labeled target MRI that serves as an atlas. The inverse of the
recovered transformation is used to warp the atlas labels back onto the
subject's MRI, thus achieving segmentation. ![]() has been shown to
successfully segment basal ganglia structures [1] but it has
not been able to segment cortical structures satisfactorily (voxel-based
overlap indexes have been typically around 40%). This is due to the fact
that the deformation field estimated by
has been shown to
successfully segment basal ganglia structures [1] but it has
not been able to segment cortical structures satisfactorily (voxel-based
overlap indexes have been typically around 40%). This is due to the fact
that the deformation field estimated by ![]() does not have the power to
unfold the cortex of one brain and then refold it back onto a target brain.
The deformation field is bandlimited and therefore does not have high
enough frequencies to introduce (or remove) cortical folds where needed.
does not have the power to
unfold the cortex of one brain and then refold it back onto a target brain.
The deformation field is bandlimited and therefore does not have high
enough frequencies to introduce (or remove) cortical folds where needed.
The second segmentation strategy is named ![]() (Intensity Normalized
Stereotaxic Environment for the Classification of Tissue). Complementary
to
(Intensity Normalized
Stereotaxic Environment for the Classification of Tissue). Complementary
to ![]() ,
, ![]() was designed to automatically identify different
tissue types from either single channel or multi-spectral MRI data. This
method has been shown to successfully classify grey matter (GM), white
matter (WM), and cerebral spinal fluid (CSF) and has been applied to the
problem of automatically segmenting multiple sclerosis (MS) lesions from a
large number of subjects in the context of a third phase clinical trial
[8]. While classification techniques such as
was designed to automatically identify different
tissue types from either single channel or multi-spectral MRI data. This
method has been shown to successfully classify grey matter (GM), white
matter (WM), and cerebral spinal fluid (CSF) and has been applied to the
problem of automatically segmenting multiple sclerosis (MS) lesions from a
large number of subjects in the context of a third phase clinical trial
[8]. While classification techniques such as ![]() are
able to separate GM, WM and CSF and in so doing, extract fine detail from
the MRI volume, they cannot differentiate between two adjacent structures
with the same tissue type.
are
able to separate GM, WM and CSF and in so doing, extract fine detail from
the MRI volume, they cannot differentiate between two adjacent structures
with the same tissue type.
By merging the complementary information from ![]() 's non-linear
deformation with the output of
's non-linear
deformation with the output of ![]() 's classification technique, it is
possible to accurately identify specific cortical structures from a
subject's MRI.
's classification technique, it is
possible to accurately identify specific cortical structures from a
subject's MRI.
The ![]() algorithm deforms one MRI volume to match another, previously
labelled, MRI volume. It builds up the 3D non-linear deformation field in
a local fashion, fitting spherical neighbourhoods in sequence. Each local
neighbourhood in one volume is translated to achieve an optimal match
within the other volume. Neighbourhoods are arranged in a 3D grid to fill
the volume and each one moves within a range defined by the grid-spacing.
The algorithm is applied in a multi-scale hierarchy. At each step, the
image volumes are pre-convolved with a 3D Gaussian kernel where blurring
and neighbourhood size are reduced at after each stage. Initial fits are
obtained rapidly since at lower scales, only gross distortions are
considered, but later iterations at finer scales accommodate local
differences at the price of increasing computational burden. Segmentation
is achieved by transforming labels from the second volume onto the first
volume, via the spatial mapping of the 3D deformation field.
algorithm deforms one MRI volume to match another, previously
labelled, MRI volume. It builds up the 3D non-linear deformation field in
a local fashion, fitting spherical neighbourhoods in sequence. Each local
neighbourhood in one volume is translated to achieve an optimal match
within the other volume. Neighbourhoods are arranged in a 3D grid to fill
the volume and each one moves within a range defined by the grid-spacing.
The algorithm is applied in a multi-scale hierarchy. At each step, the
image volumes are pre-convolved with a 3D Gaussian kernel where blurring
and neighbourhood size are reduced at after each stage. Initial fits are
obtained rapidly since at lower scales, only gross distortions are
considered, but later iterations at finer scales accommodate local
differences at the price of increasing computational burden. Segmentation
is achieved by transforming labels from the second volume onto the first
volume, via the spatial mapping of the 3D deformation field.
The main advantage of this type of segmentation methodology is that it results in atlas-independent segmentation since structure identification is simply a by-product of non-linear registration. The anatomical labels defined on the target volume are not used in any way to determine the match between data and model. Therefore, any atlas defined on the target MRI brain volume can be used for segmentation, thereby allowing for the co-existence of multiple atlases, each of which is simultaneously mappable to the native MR image volume without the CPU-expensive recalculation of the non-linear spatial transformation required for registration.
![]() is a production pipeline aimed at the fully automatic analysis of
large volumes of MRI data [7]. It is based on stereotaxic
registration which maps the MRI data into a standardized, Talairach
coordinate space using a 9-parameter linear transformation
[3] that allows, among others, the use of spatial masks and
priors, facilitates longitudinal and between-group analysis, and provides a
means for voxel-based statistical analysis.
is a production pipeline aimed at the fully automatic analysis of
large volumes of MRI data [7]. It is based on stereotaxic
registration which maps the MRI data into a standardized, Talairach
coordinate space using a 9-parameter linear transformation
[3] that allows, among others, the use of spatial masks and
priors, facilitates longitudinal and between-group analysis, and provides a
means for voxel-based statistical analysis. ![]() assigns each voxel in
stereotaxic space a tissue class label (e.g., WM, GM, CSF) using an
artificial neural network (ANN) classifier. The classifier relies on input
from one or more MRI volumes (e.g., T1-, T2-, and PD-weighted scans),
which have previously been registered with and resampled into stereotaxic
space. The ANN is trained for each individual subject scan using a fixed
set of stereotaxic training locations for each tissue class, derived from
SPAMs of WM, GM, and CSF [5].
assigns each voxel in
stereotaxic space a tissue class label (e.g., WM, GM, CSF) using an
artificial neural network (ANN) classifier. The classifier relies on input
from one or more MRI volumes (e.g., T1-, T2-, and PD-weighted scans),
which have previously been registered with and resampled into stereotaxic
space. The ANN is trained for each individual subject scan using a fixed
set of stereotaxic training locations for each tissue class, derived from
SPAMs of WM, GM, and CSF [5].
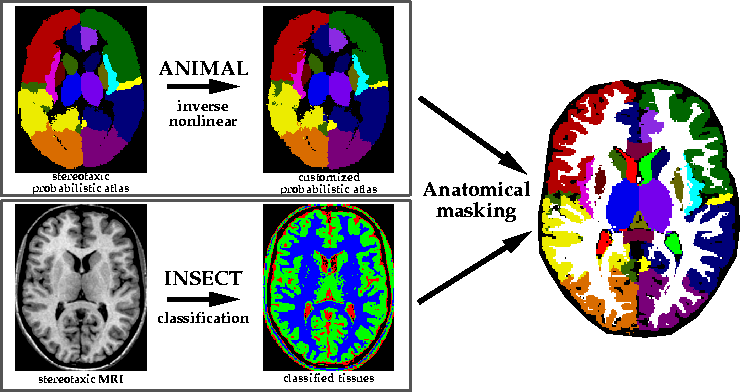
The complementary information from ![]() 's non-linear deformation (i.e.,
low resolution region identification) was merged with the output of
's non-linear deformation (i.e.,
low resolution region identification) was merged with the output of
![]() 's classification technique (i.e., voxel class labels) in order to
accurately identify specific structures in a subjects MRI (see
Fig. 1). In short, the grey matter (extracted by
's classification technique (i.e., voxel class labels) in order to
accurately identify specific structures in a subjects MRI (see
Fig. 1). In short, the grey matter (extracted by ![]() )
is masked by a structural region mask (defined by
)
is masked by a structural region mask (defined by ![]() ) in order to
define specific cortical brain structures. Similarly, CSF-labeled voxels
are masked with ventricular masks to identify the lateral, third and forth
ventricles.
) in order to
define specific cortical brain structures. Similarly, CSF-labeled voxels
are masked with ventricular masks to identify the lateral, third and forth
ventricles.
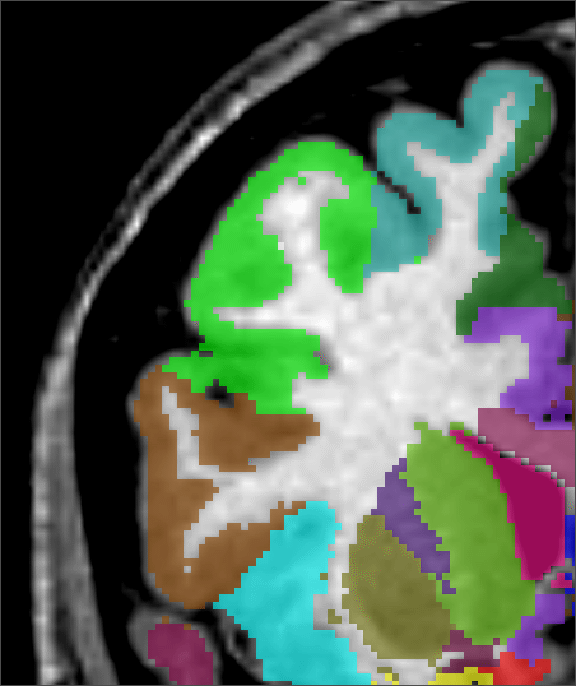
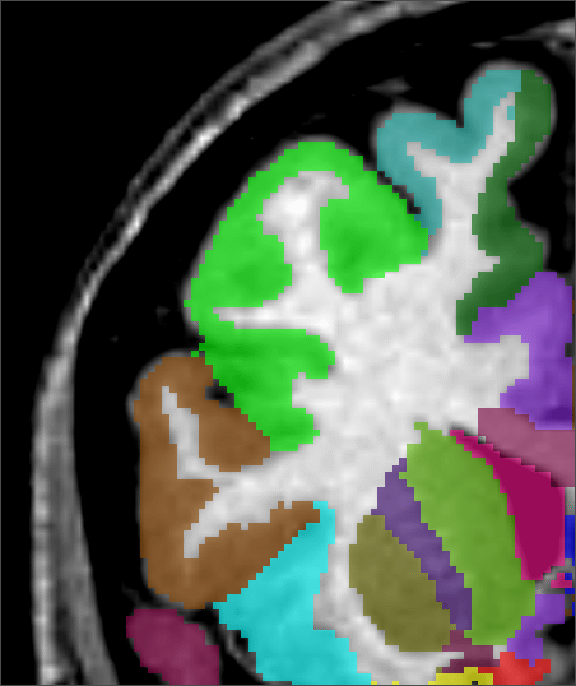
Figure 2 shows a comparison of an ![]() -only segmentation, an
-only segmentation, an
![]() +
+![]() segmentation and a manual segmentation. Not only is the
segmentation and a manual segmentation. Not only is the
![]() +
+![]() segmentation improved at the cortex, where some
grey-matter regions were missed with the standard
segmentation improved at the cortex, where some
grey-matter regions were missed with the standard ![]() technique, the
segmentation of the lateral ventricles is much better as well (on other
planes). Where the
technique, the
segmentation of the lateral ventricles is much better as well (on other
planes). Where the ![]() technique overestimated the size of the
ventricle, the
technique overestimated the size of the
ventricle, the ![]() +
+![]() is in complete agreement with the MRI
anatomy and with the expert's labelling.
is in complete agreement with the MRI
anatomy and with the expert's labelling.
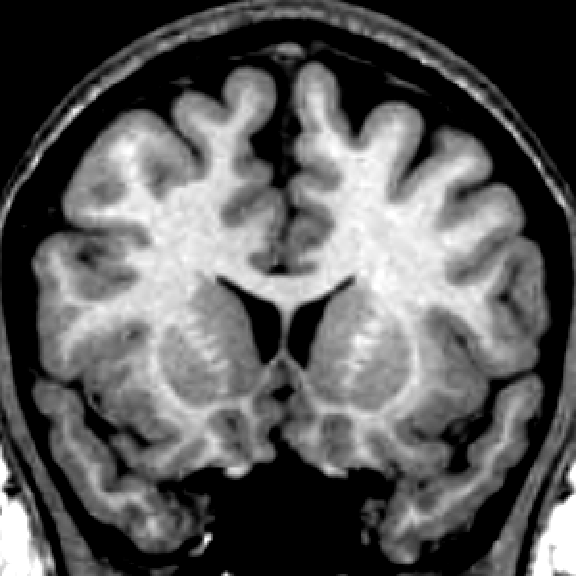 |
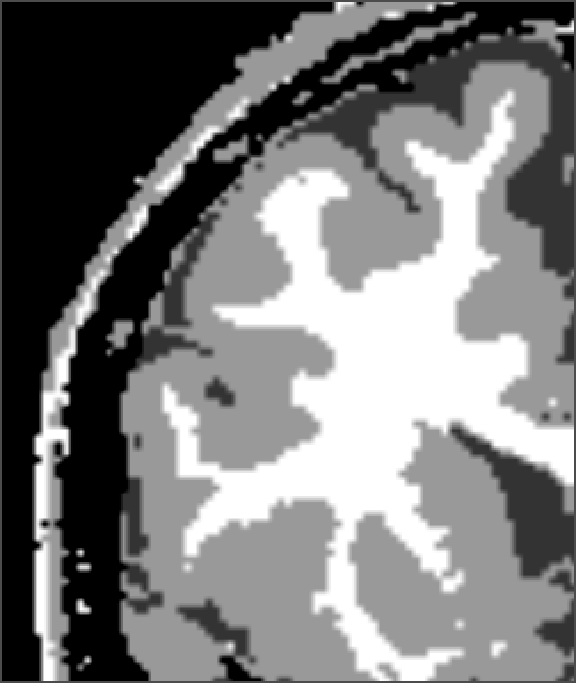 |
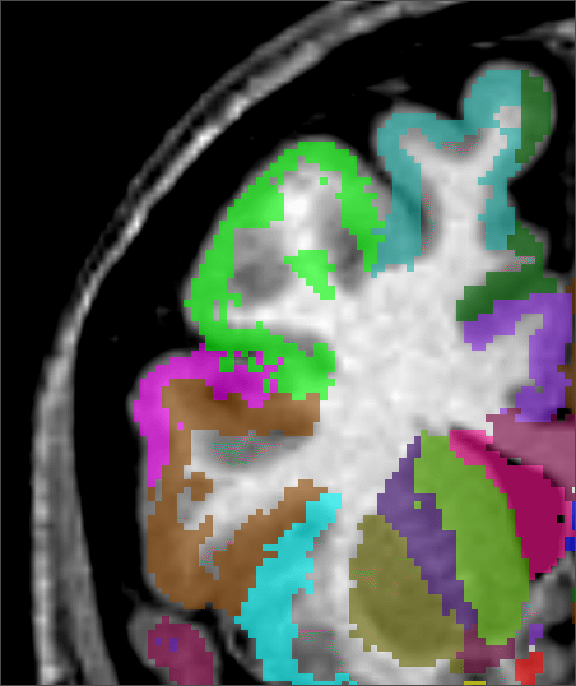 |
 |
 |
| input MRI | manual | |||
| classification | segmentation | segmentation | segmentation |
The ![]() +
+![]() method has been applied to 150 data sets acquired as
part of the HBMP-funded International Consortium for Brain Mapping (ICBM)
project [6] where the goal is to estimate structure volumes
[2] and to create a SPAM (statistical probability anatomy
map) atlas [4] that quantifies the normal anatomical
variability of both shape and position for the different regions of the
normal brain.
method has been applied to 150 data sets acquired as
part of the HBMP-funded International Consortium for Brain Mapping (ICBM)
project [6] where the goal is to estimate structure volumes
[2] and to create a SPAM (statistical probability anatomy
map) atlas [4] that quantifies the normal anatomical
variability of both shape and position for the different regions of the
normal brain.
Since ![]() yields high resolution structure information, it is no
longer necessary to run
yields high resolution structure information, it is no
longer necessary to run ![]() to fine resolutions, thus providing a
considerable improvement in speed. While the procedure described here uses
two algorithms that were developed at the Montreal Neurological Institute,
the new improved segmentation method is not dependent on these particular
methods. In fact, any classification method that differentiates tissue
types and any non-linear registration method may be merged to maximize the
complementary information of both techniques. The method presented is
completely automatic, therefore fully objective and applicable to large
ensembles of brain volumes.
to fine resolutions, thus providing a
considerable improvement in speed. While the procedure described here uses
two algorithms that were developed at the Montreal Neurological Institute,
the new improved segmentation method is not dependent on these particular
methods. In fact, any classification method that differentiates tissue
types and any non-linear registration method may be merged to maximize the
complementary information of both techniques. The method presented is
completely automatic, therefore fully objective and applicable to large
ensembles of brain volumes.
The authors would like to express their appreciation for support from the Human Frontier Science Project Organization, the Canadian Medical Research Council (SP-30), the McDonnell-Pew Cognitive Neuroscience Center Program, the U.S. Human Brain Map Project (HBMP), NIMH and NIDA. This work forms part of a continuing project of the HBMP-funded International Consortium for Brain Mapping (ICBM) to develop a probabilistic atlas of human neuroanatomy. We also wish to acknowledge the manual anatomical labelling completed by Colin Holmes and Noor Kabani.