Abstract
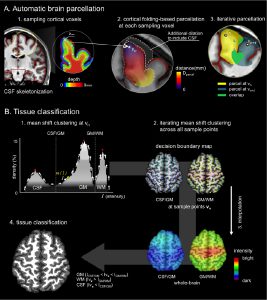
Accurate tissue classification is a crucial prerequisite to MRI morphometry. Automated methods based on intensity histograms constructed from the entire volume are challenged by regional intensity variations due to local radiofrequency artifacts as well as disparities in tissue composition, laminar architecture and folding patterns. Current work proposes a novel anatomy-driven method in which parcels conforming cortical folding were regionally extracted from the brain. Each parcel is subsequently classified using nonparametric mean shift clustering. Evaluation was carried out on manually labeled images from two datasets acquired at 3.0 Tesla (n = 15) and 1.5 Tesla (n = 20). In both datasets, we observed high tissue classification accuracy of the proposed method (Dice index >97.6% at 3.0 Tesla, and >89.2% at 1.5 Tesla). Moreover, our method consistently outperformed state-of-the-art classification routines available in SPM8 and FSL-FAST, as well as a recently proposed local classifier that partitions the brain into cubes. Contour-based analyses localized more accurate white matter-gray matter (GM) interface classification of the proposed framework compared to the other algorithms, particularly in central and occipital cortices that generally display bright GM due to their highly degree of myelination. Excellent accuracy was maintained, even in the absence of correction for intensity inhomogeneity. The presented anatomy-driven local classification algorithm may significantly improve cortical boundary definition, with possible benefits for morphometric inference and biomarker discovery.