When DTI images are obtained by EPI sequence, they inherently involve various noises, such as eddy current distortion, head motion and B0 inhomogeneity distortion. These noises significantly degrade the results of DTI estimates or DTI tractography if you obtained the data in higher spatial or angular resolution. Such image distortions cannot be properly corrected even using a non-linear registration method. The script, dti_preprocess, does correct them and calculate diffution tensor estimates (FA, MD, V1-3, L1-3 etc.) using FSL software. You can use the results for any tractographic software that handles ANALYZE7.5 or NIFTI format images (FDT, DtiStudio, dTV etc) or for DTI-based statistics (e.g. TBSS).
To use this script, you need a computer in which FSL is already installed, and data (DTI data; fieldmap data: fieldmap and magnitude; MPG vector information :bvec and bval; and dwell time and TE for the DTI). The DTI and fieldmap data must be in NIFTI or ANALYZE7.5 format. At the first step, the script does rescale the original data in higher resolution (by default, x0.6 of the original x, y dimension). Next, it corrects the eddy current distortion and/or motion by 12-DOF linear registration (this is same as used in eddy_correct in FSL), and then the B0 distortion by using B0 fieldmap data (magnitude and phase) and fugue. It then correct the b-vector by using rotations deteremined from motion estimates. Finally, it calculates DTI estimates by dtifit. The image reslicing does not get applied at the same time as the estimation of motion and/or eddy-current distortion; instead the reslice gets applied simultaneously with the application of B0 unwarping in order to minimize interpolation related image blurring.
Installation
1. Download dti_preprocess from here (current version 2.0).
2. Log in as root, and save the downloaded dti_preprocess in the directory whereever search path is set (e.g. /usr/bin/).
3. Type
chmod +x /usr/bin/dti_preprocessThis makes it possible for every FSL user to run dti_process. Just typing
dti_preprocessor
dti_preprocess -helpin the terminal will show the usage or help page.
Example usage 1 (test calculation)
dti_preprocess -k dti.img -f fm.img -m mag.img -t 0.76 -e 100This performs test dewarping only for b=0 image. The flag -d is followed by 4D DTI data (dwi.img, whose first volume must be b=0 image), -f: by fieldmap image (fm.img in units of rad/sec), -m: by fieldmap magnitude image (mag.img), -t: by dwell time (0.76 ms) for DTI, and -e: by TE (100 ms) for DTI. The calculation finishes in a relatively shorter time, so that you can check whether all the parameters you use (dwell time, unwarp direction, reslicing resolution, etc.) are optimal before doing full calculation (see Example usage 2). Note that the fieldmap image (fm.img) must be unwrapped in phase and have units of rad/sec. This usually requires site/scanner/sequence specific preprocessing (see prelude/fugue in FSL web), but Siemens users may want to use ph2fm_siemens. To see the results of dti_preprocess, point your browser at the report.html in the output directory (default ourputdir= ./dti).
Example usage 2 (full calculation)
dti_preprocess -k dti.img -f fm.img -m mag.img -t 0.76 -e 100 -b bvals -r bvecsThis performs full calculation of dti data, and it takes longer time. You need additional flags -b and -r, each followed by text file, bvals and bvecs, which contains information of b-values or MPG vectors, respectively. These text files must be formatted as described below. The final outputs (unwarped data, DTI estimates, signal loss image etc.) will be all stored in the output directory (default = ./dti). This output directory can be directly used as an input for the subsequent Baysian estimation of the local diffusion parameters (bedpostx in FSL). The signal loss image (nodif_sigloss) can also be used as a weight image when registering images between DTI and other modalities (e.g. fMRI image or structural T1 image).
The text files, bvecs and bvals, must be formatted as follows:
bvals: the order of entries in this file must match the order of volumes in the DTI data and entries in the gradient directions text file. The format is:
b_1 b_2 b_3 ... b_n
bvecs: the order of entries in this file must also match the order of volumes in the DTI data series. The format is:
x_1 x_2 x_3 ... x_n
y_1 y_2 y_3 ... y_n
z_1 z_2 z_3 ... z_n
For volumes in which there was no diffusion weighting, the entry should still be present as x=0,y=0,z=0.
Other options include
-u [x, x-, y, y-, z, or z-] : unwarp direction (default: y-)
-s [num] : %signal loss threshold for B0 unwarping (default: 10)
-v [num] : reslice resolution in mm (default: no reslicing)
-K [img] : mask file for dti image (by default, this is calculated by bet)
-M [img] : mask file for fieldmap magnitude image (by default, this is calculated by bet)
-d [dir] : directory to save outputs (default: ./dti)
-F : Use the actual directory name given (i.e. do not add + to make a new directory)
-R [num] : reference volume (default : 0th volume)
-4 : 3-step registration in mcflirt (slow but more accurate, default is 3-step)
-l : use flirt instead of mcflirt (much slower but more accurate)
-n : do not register between fieldmap and dti data
-B : correct b-vector file using rotation parameters (output bvecsecc)
-U : do not mask DTI images
OutputsThe outputs of dti_preprocess includes:
bvecs, bvals: vector file and b-value file. dti_FA, dti_L1, dti_L2, dti_L3, dti_V1, dti_V2, dti_V3, dti_MD, dti_MO, dti_S0, dti_V1, dti_V2, dti_V3, dti_sse: outputs of dtifit. dti_preprocess: a directory including all the intermediate files. nodif, nodif_brain, nodif_brain_mask: non-diffusion-weighted (b=0) images. nodif_sigloss: signal loss images of non-diffusion-weighted (b=0) image. This image might be useful for weighting registration. report.html: report of dti_preprocess in html format.Example results
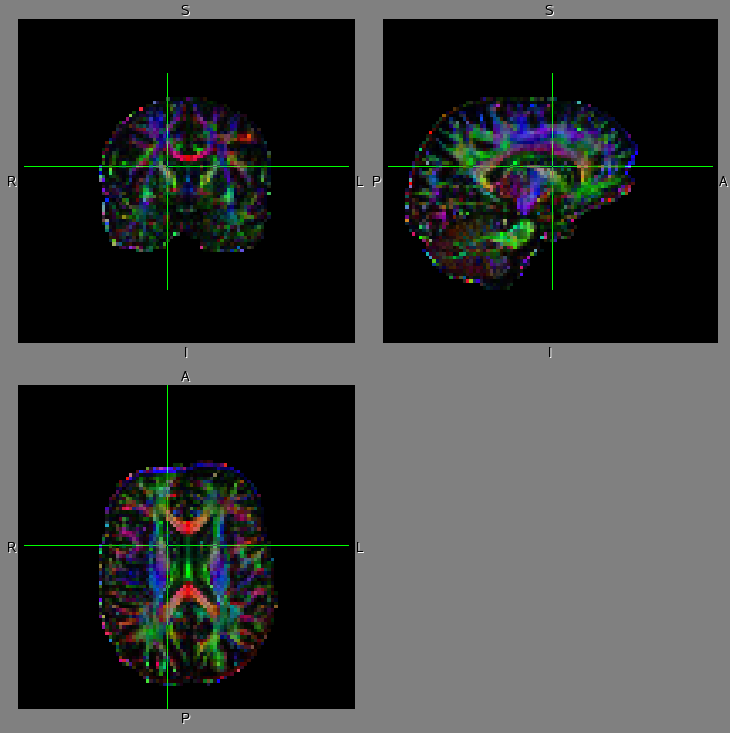
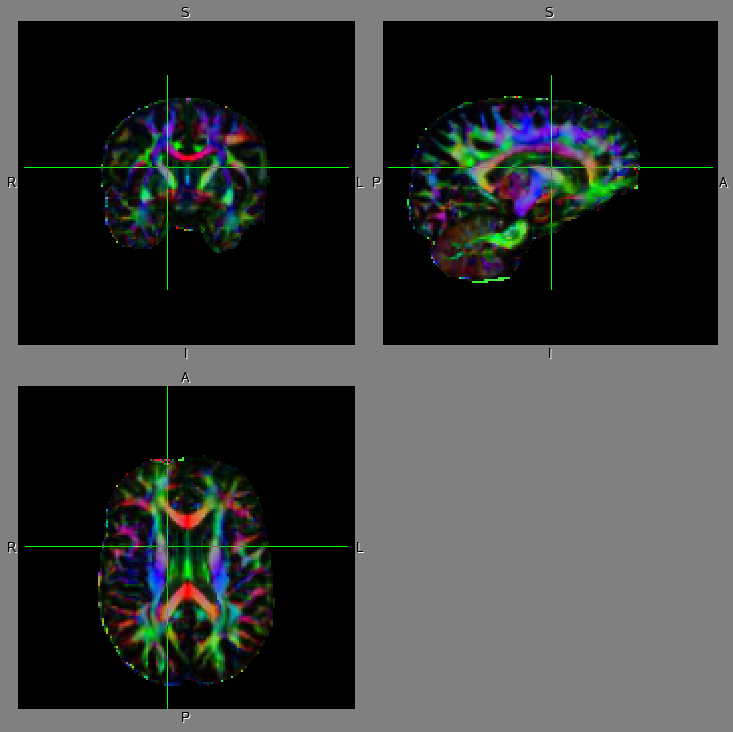
The DTI data was obtained by Siemens 1.5T Sonata scanner at MNI (spatial resolution: 2.5mm-cubic voxel; the number of MPG direction: 60; phase direction: AP). Left shows the dti estimates, FA: fractional anisotropy and V1: the 1st eigenvector of diffusion tensor before dti_preprocess and right shows those after dti_preprocess. The V1 is displayed in RGB color (R: right-left; G: anterior-posterior; B: superior-inferior) overlaid on FA image (dark: low FA value; bright: high FA value). Note that the distortion in the fronto-polar region (which is caused by the frontal sinus and is often significant when you obtained DTI data in the phase direction of AP) is properly corrected.