



Next: Conclusions and Discussion
Up: Results
Previous: 3D Results - Neck
Solution of the fluid PDE in the spatial domain using finite differencing and relaxation is slow. We restricted the upper limit of the number of SOR iterations within each timestep to 4000![[*]](/icons/latex2html/foot_motif.gif) in the 2D case and to 50 in the 3D case. This provided a compromise between speed and accurate solution of the PDE, since within each timestep computation of velocities is approximate and is improved on in the next timestep. For images sized
in the 2D case and to 50 in the 3D case. This provided a compromise between speed and accurate solution of the PDE, since within each timestep computation of velocities is approximate and is improved on in the next timestep. For images sized
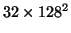 ,
50 SOR iterations took 2 minutes 36 seconds for the constant-viscosity fluid on a Sun UltraSPARC - equivalent to 416 minutes for 4000 iterations for a
,
50 SOR iterations took 2 minutes 36 seconds for the constant-viscosity fluid on a Sun UltraSPARC - equivalent to 416 minutes for 4000 iterations for a
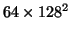 image.
[14] have implemented full-multigrid (FMG) solution of the same PDE applied to the displacement field for elastic registration; they estimate 6,592 Jacobi relaxation iterations for a
image.
[14] have implemented full-multigrid (FMG) solution of the same PDE applied to the displacement field for elastic registration; they estimate 6,592 Jacobi relaxation iterations for a
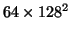 grid. For a grid sized
grid. For a grid sized
 ,
SOR requires
,
SOR requires
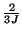 the number of iterations compared to Jacobi relaxation [15]. [14] show that two 3D FMG cycles are sufficient for a
the number of iterations compared to Jacobi relaxation [15]. [14] show that two 3D FMG cycles are sufficient for a
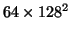 grid to solve the elasticity PDE per iteration, giving 20 minutes per iteration. Hence we estimate that replacing SOR with FMG would speed the fluid registration by a factor of 20.
grid to solve the elasticity PDE per iteration, giving 20 minutes per iteration. Hence we estimate that replacing SOR with FMG would speed the fluid registration by a factor of 20.
MF1 additionally multiplies each pixel by (6) once per timestep, a minimal overhead compared to the
SOR iterations. MF2 provides a considerable gain in speed over UF, depending on the volume percentage of rigid bodies
(pixels whose velocities are not computed).
The percentage of passive regions in the image is equal to the percentage speed-up in the SOR solution. For MF3, extra finite differencing computations are added to the SOR due to the extra terms in (7); we timed 3 minutes 47 seconds for 50 SOR iterations for images sized
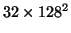 .
.




Next: Conclusions and Discussion
Up: Results
Previous: 3D Results - Neck
Hava LESTER
1999-03-24
![]() .
.