



Next: Computational Time
Up: Results
Previous: Pre/post-operative Head Images, Rigid
3-dimensional versions of UF and MF3 were compared on MRI neck volumes with the vertebrae defined as weakly deformable.
The original images were acquired at the Hammersmith hospital![[*]](/icons/latex2html/foot_motif.gif) .
Two full-3D neck volumes were provided, neckD
and neckI. NeckI was of the chin down and neckD was with the head flexed backwards within the confines of the scanner bore.
Both were of the same subject.
Since the imaging field of view had been the spinal column specifically, the volumes did not extend to include the whole neck laterally.
The images exhibited a strong intensity ramp, with high values at the back of the neck and total signal loss in the face. We pre-processed the images to give a more uniform range of intensities in the anterior-posterior direction, using the scheme described by Fig. 7. Due to memory capacity and time constraints, it was not feasible to apply the fluid registrations directly to the full-resolution data sets; hence they were downsampled, by blurring with a Gaussian of standard deviation
.
Two full-3D neck volumes were provided, neckD
and neckI. NeckI was of the chin down and neckD was with the head flexed backwards within the confines of the scanner bore.
Both were of the same subject.
Since the imaging field of view had been the spinal column specifically, the volumes did not extend to include the whole neck laterally.
The images exhibited a strong intensity ramp, with high values at the back of the neck and total signal loss in the face. We pre-processed the images to give a more uniform range of intensities in the anterior-posterior direction, using the scheme described by Fig. 7. Due to memory capacity and time constraints, it was not feasible to apply the fluid registrations directly to the full-resolution data sets; hence they were downsampled, by blurring with a Gaussian of standard deviation
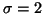 and storing alternate pixels (Fig. 7, right). Since the Gaussian blurring for downsampling, and intensity gradient calculations during registration, were performed in the Fourier domain and required image dimensions of powers of two, we used zero padding to give full resolution dimensions
and storing alternate pixels (Fig. 7, right). Since the Gaussian blurring for downsampling, and intensity gradient calculations during registration, were performed in the Fourier domain and required image dimensions of powers of two, we used zero padding to give full resolution dimensions
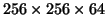 and downsampled volumes of
and downsampled volumes of
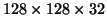 .
.
Figure:
Pre-processing stages shown on neckD. ( left-right): slice 19 of the original 3D
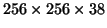 volume; after division pixelwise by the same image blurred with a Gaussian of spatial standard deviation
volume; after division pixelwise by the same image blurred with a Gaussian of spatial standard deviation
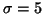 ;
masked with the aid of the automatic contouring and manual alteration tool in xdispunc; downsampled by a half
;
masked with the aid of the automatic contouring and manual alteration tool in xdispunc; downsampled by a half
![\begin{figure}\makebox[\textwidth]{
\epsfxsize=0.2\textwidth \epsfbox{Dorig19of3...
...x{Dmask19of38.ps}
\epsfxsize=0.2\textwidth \epsfbox{Dhf16of32.ps}
}
\end{figure}](img49.gif) |
We segmented the spinal vertebrae slice-by-slice from the full-resolution source volumes using the xdispunc display tools. The contrast between vertebrae and intervening tissue was variable and so segmentation was performed manually with reference to an atlas [13].
The segmentations were converted to
binary spine volumes which were then downsampled using the same process as for the necks.
Both fluid registration tests (UF, MF3)
were applied in a
six-level scale space (Gaussian blurs of spatial standard deviation 2i with
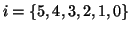 ). Within each scale level, the fluid was set to iterate through at least three timesteps, with an optional extra 100 timesteps until the stopping criterion was met, the stopping criterion being a reduction in correlation coefficient of less than 10-4.
On termination, we upsampled the displacement fields obtained from both fluid registrations of neckD to neckI and applied them to the original neckD images, to give full-resolution deformations. These are shown in Fig. 8 (top, centre).
). Within each scale level, the fluid was set to iterate through at least three timesteps, with an optional extra 100 timesteps until the stopping criterion was met, the stopping criterion being a reduction in correlation coefficient of less than 10-4.
On termination, we upsampled the displacement fields obtained from both fluid registrations of neckD to neckI and applied them to the original neckD images, to give full-resolution deformations. These are shown in Fig. 8 (top, centre).
Figure 8:
( top) Central slices of full resolution neckD ( left) and neckI ( right). ( centre-left) UF and ( centre-right) MF3 registration of neckD to neckI. ( bottom) Central slices of ( left to right:) neckD - neckI; UF neckD - neckI; MF3 fluid neckD - neckI
![\begin{figure}
\makebox[\textwidth]{
\epsfysize=0.2\textwidth \epsfbox{D32.ps}
\...
...sfbox{vDtoIfrI16.ps}
\epsfxsize=0.2\textwidth \epsfbox{blank.eps}
}
\end{figure}](img51.gif) |
We applied the transformation fields produced by
both fluid tests to the intial spine volumes segmented from neckD;
the results of the volume-rendered spines are shown from two angles in
Fig. 9.
The UF registration shows an extension of the upper two vertebrae of the spines on comparison to the original segmentation from neckD (Fig. 9 far right).
Figure 9:
( left-right) original spineD;
spineD after UF registration of neckD to neckI;
spineD after MF3 registration of neckD to neckI with vertebrae weakly deformable; ( far right): the upper three vertebrae after ( left) UF and ( right ) MF3 registration.
The 3D images were volume-rendered using the Analyze package
![\begin{figure}\makebox[\textwidth]{
\epsfxsize=0.10\textwidth \epsfbox{vgs_Dspin...
....15\textwidth]{
\epsfysize=0.182\textwidth \epsfbox{var_bit.ps}
}
}
\end{figure}](img52.gif) |
Figure 10
shows logs of the Laplacian and elasticity energies as local deformation metrics computed from the displacements of both registrations of neckD
to neckI.
The deformation metrics clearly show dark patches in the vertebrae in MF3 indicating low distortion: compare Fig. 10 (left) with those of the UF (Fig. 10 right).
Figure 10:
Central slices of the deformation metric images, after registration of neckD to neckI ( left) log of Laplacian, UF; ( centre left) log of elasticity energy, UF;
( centre right and far right) the equivalent for MF3
![\begin{figure}\makebox[\textwidth]{
\epsfxsize=0.18\textwidth \epsfbox{fDtoIlLa1...
...{vDtoIlLa16.ps}
\epsfxsize=0.18\textwidth \epsfbox{vDtoIlel16.ps}
}
\end{figure}](img53.gif) |




Next: Computational Time
Up: Results
Previous: Pre/post-operative Head Images, Rigid
Hava LESTER
1999-03-24
![]() ). Within each scale level, the fluid was set to iterate through at least three timesteps, with an optional extra 100 timesteps until the stopping criterion was met, the stopping criterion being a reduction in correlation coefficient of less than 10-4.
On termination, we upsampled the displacement fields obtained from both fluid registrations of neckD to neckI and applied them to the original neckD images, to give full-resolution deformations. These are shown in Fig. 8 (top, centre).
). Within each scale level, the fluid was set to iterate through at least three timesteps, with an optional extra 100 timesteps until the stopping criterion was met, the stopping criterion being a reduction in correlation coefficient of less than 10-4.
On termination, we upsampled the displacement fields obtained from both fluid registrations of neckD to neckI and applied them to the original neckD images, to give full-resolution deformations. These are shown in Fig. 8 (top, centre).