


Study of the Genetic Encoding of the Central Sulcus Shape Using Principal
Component Analysis
G Le Goualher , A.M. Argenti
, A.M. Argenti 
 and A.C. Evans
and A.C. Evans
 McConnell Brain Imaging Centre, Montréal Neurological
Institute,
McConnell Brain Imaging Centre, Montréal Neurological
Institute,
McGill University, Montréal, Canada ;
 Unité de Recherche d'épidemiologie génétique,
INSERM,Unité 155, Université Paris VII, Jussieu, Paris
Unité de Recherche d'épidemiologie génétique,
INSERM,Unité 155, Université Paris VII, Jussieu, Paris
Principal Component Analysis (PCA) allows the description of shape
variability with a restricted number of parameters. These parameters (or
modes) can be used to quantify the difference between two shapes through
the computation of a modal distance. A statistical test can then
be applied to this set of measurements in order to detect a significant
difference between two groups of subjects. We have applied this
methodology to highlight the genetic encoding of the shape of an
anatomical structure. To underline genetic constraint, we have studied
if central sulci were more similar within pairs of monozygotic twins than
between independent subjects. This comparison was performed using a Mantel
Permutation Test.
We had at our disposal 20 tri-dimensional MRI volumes corresponding to 10
pairs of monozygotic twins (average age : 34.6 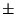 9.85; 8 males, 12
females). We have also included in our analysis 20 additional
tri-dimensional MRIs corresponding to 20 subjects (average age : 29.6
9.85; 8 males, 12
females). We have also included in our analysis 20 additional
tri-dimensional MRIs corresponding to 20 subjects (average age : 29.6 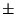 6.22; 8 males, 12 females) selected from a
database of 150 volunteers corresponding to normal young
subjects acquired as part of the International Consortium for Brain Mapping
(ICBM) project.
6.22; 8 males, 12 females) selected from a
database of 150 volunteers corresponding to normal young
subjects acquired as part of the International Consortium for Brain Mapping
(ICBM) project.
We have previously developed a method (referred to as an
Active Ribbon) which models the median surface of a sulcus by a
parametric surface [1]. Principal Component Analysis (PCA) can be applied to a
collection of such numerical representation of shapes. This eigenanalysis
technique quantifies shape in term of deformation modes which
represent unique and natural coordinates for shape comparisons. In the
modal space, modal distance quantifies the observed difference
between two shapes. In the eigen vector space, the modal distance between two samples
is defined as :
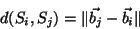 |
(1) |
The closer this norm is to zero, the more similar the two samples are.
If a genetic encoding exists, one can argue that the minimum sulcal shape
distance
dmin(Si,Sj) will systematically occur within a pair of
twins :
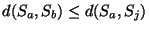 ,
,
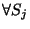 ,
if Sa and Sb is
a pair of twins.
However, this assumption may not be true even if a genetic encoding exists simply by
chance. A statistical test is therefore needed to show
that the intra-pair modal distances are statistically inferior to
inter-pair modal distances.
,
if Sa and Sb is
a pair of twins.
However, this assumption may not be true even if a genetic encoding exists simply by
chance. A statistical test is therefore needed to show
that the intra-pair modal distances are statistically inferior to
inter-pair modal distances.
The statistical we have used is the Mantel Permutation
Test [2]. The
Mantel Permutation Test requires no knowledge of population distribution
(by opposition the classical student t-test is applied for the comparison
of two populations each of them following a normal distribution; moreover
to apply a student t-test the variance of the two populations must be
similar) and makes meaningful use of all available data [3].
Our study highlights genetic
constraints on the shape of the central sulcus. We found from 10 pairs of
monozygotic twins that the intra-pair modal distances of the central
sulcus were significantly inferior from inter-pair modal distances for
the left central sulcus (z = -3.3179 ; p < 0.0005 ) as well as for the
right central sulcus (z = -2.5030; p < 0.006). However, since the
magnitude of the z-values is not large, it appears that environmental
factors play a non-negligible role in the definition of the central
sulcus shape.
- 1
-
G. LE GOUALHER, C. BARILLOT, and Y. BIZAIS.
Modeling cortical sulci using active ribbons.
International Journal of Pattern Recognition and Artificial
Intelligence, 11(8):1295-1315, 1997.
- 2
-
N. MANTEL.
The detection of disease clustering and a generalized regression
approach.
Cancer Research, 27:209-220, 1967.
- 3
-
K.J. WORSLEY, A.C. EVANS, S.C. STROTHER, and
J.L. TYLER.
A Linear Spatial Correlation Model, with Applications to
Positron Emission Tomography.
Journal of the American Statistical Association,
86(413):55-67, March 1991.
 Supported by U.S. Human Brain Map Project and the
International Consortium for Brain Mapping.
The authors are grateful to Michel Duyme of Unité de Recherche d'épidemiologie génétique,
INSERM,Unité 155, Université Paris VII, Jussieu, Paris for
providing them with the twins data.
Supported by U.S. Human Brain Map Project and the
International Consortium for Brain Mapping.
The authors are grateful to Michel Duyme of Unité de Recherche d'épidemiologie génétique,
INSERM,Unité 155, Université Paris VII, Jussieu, Paris for
providing them with the twins data.
This document was generated using the
LaTeX2HTML translator Version 98.1p1 release (March 2nd, 1998)
Copyright © 1993, 1994, 1995, 1996, 1997,
Nikos Drakos,
Computer Based Learning Unit, University of Leeds.
The command line arguments were:
latex2html -split 0 genetic.tex genetic.html.
The translation was initiated by Georges Le Goualher on 1999-01-13



Georges Le Goualher
1999-01-13
![]() , A.M. Argenti
, A.M. Argenti ![]()
![]() and A.C. Evans
and A.C. Evans![]()
![]() McConnell Brain Imaging Centre, Montréal Neurological
Institute,
McConnell Brain Imaging Centre, Montréal Neurological
Institute,![]() Unité de Recherche d'épidemiologie génétique,
INSERM,Unité 155, Université Paris VII, Jussieu, Paris
Unité de Recherche d'épidemiologie génétique,
INSERM,Unité 155, Université Paris VII, Jussieu, Paris