For an introduction to IGNS, see IGNS and Ultrasound in IGNS.
The goal of this work is to update the patient's pre-operative MR images based
on US images acquired intra-operatively, given the presence of non-linear
brain deformations. Our strategy is to first acquire pre-operative patient MRI
and store the result as a 3D volume. During the surgical procedure, a series
of US images are acquired. The specific aims of this work are: (i) to compute
an initial linear transformation that maps the position and orientation of the
US images into the coordinate space defined by the pre-operative
MR volume, (ii) to construct a full 3D composite US volume from the images
acquired and (iii) to compute the non-linear deformation field from the US
volume to the corresponding MR volume in order to correct for non-linear brain
deformations as well as errors in the linear registration stage. The resulting
deformation field will be used to provide the surgeon with an update of the
patient's MR images during the procedure.
Pre-operative patient MRI are acquired and stored as a 3D volume in MINC format, a publicly available medical image file format
developed at the Montreal
Neurological Institute that was designed as a multi-modal, N-dimensional,
cross-platform format. In order to perform comparisons between the two data
sets, US images acquired intra-operatively must then be mapped to the same
space as the corresponding pre-operative MRI. The transformations required to
perform the mapping are computed by making use of a Polaris tracking system
(Northern Digital Inc.) using a procedure described in US in IGNS . As US images are acquired during surgery,
the final transformation can be used to extract a corresponding oblique slice
from the MRI volume, permitting the simultaneous display and comparison of the
pre-operative MRI and intra-operative US.
Once the sequence of acquired US images are stored in the appropriate
coordinate space, a composite 3D US is created by superimposing and averaging
the slices into a full 3D volume (originally zero-valued) in the same
coordinate frame as the MR volume acquired earlier. The methodology chosen
uses a nearest neighbour approach to place each intensity pixel into the
nearest voxel in the volume image, in a strategy similar to that in [2].
Because the images are stored in MINC format, tools are
available to average the slices in the areas where they intersect. A blurring
operator is applied to the volume to remove the effects of acoustic artifacts
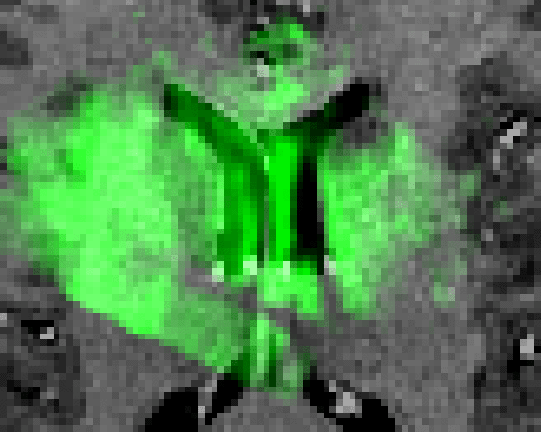
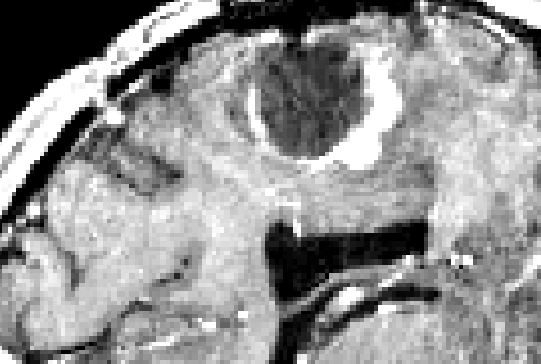
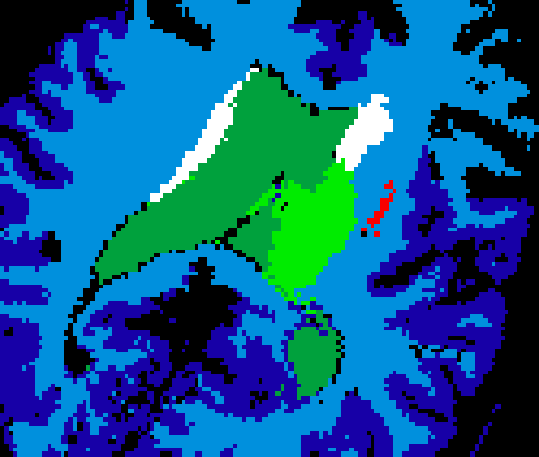
such as speckle noise. An example of an US volume superimposed onto a
patient's MRI can be seen in Figure 1.



In previous work, we have developed a versatile 3D volume registration and segmentation package, termed ANIMAL (Automatic Non-linear Image Matching and Anatomical Labeling)[1,3]. In this project, ANIMAL is primarily used to compute the non-linear spatial transformation required to map intra-operative US to pre-operative MRI. The procedure for estimating this transformation is to compute dense field of 3D deformation vectors, mapping each voxel of one image volume to match those in a target image volume. The algorithm is described in ANIMAL. However, ANIMAL requires similar features in the source and target volumes in order to perform either cross-correlation or optical flow computations between the two data sets. Since US and MRI have very different characteristics, we have decided to generate pseudo US images - images whose appearance closely matches the predicted appearance of real US images that will be acquired during surgery - from data derived from the pre-operative MRI (The current strategy does not take acoustic properties or physics into account.). In this manner, the ANIMAL routine can be used to correlate the pseudo US to the real, intra-operative US images acquired.
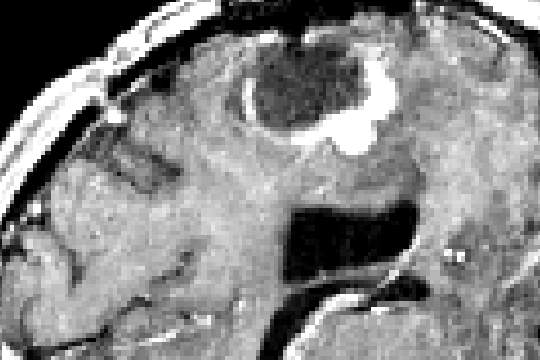
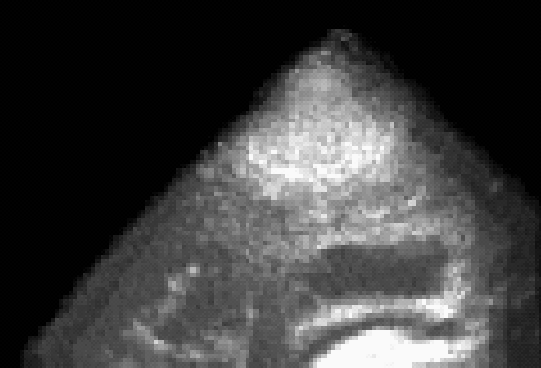
To compute the pseudo US volumes, the ANIMAL segmentation package is used
to segment major brain structures from the MRI volume. The segmentation is
then further refined to create a volume that includes only those structures
that are clearly visible in US. The resulting volume of anatomical structures
is then submitted to a radial gradient operator in order to generate gradient
magnitude data that reflects the appearance (in terms of intensity values) of
acoustic boundaries visible in the US image. In future work, other structures
prevalent in the US image (such as the cerebral falx) will be added to the
pseudo US images, and more realistic physical simulations will be developed.
However, the current technique allows for a proof-of-principal to be
demonstrated here. For the time being, pre-operative segmentation of
pathologies is performed manually (The automatic segmentation of pathologies
is currently an open research topic.). Figure 2 shows an example of each of
the steps involved in creating a pseudo US image (0.5 mm^2 pixel size).

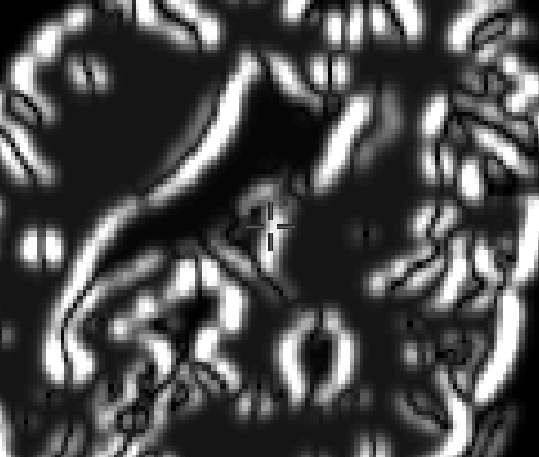
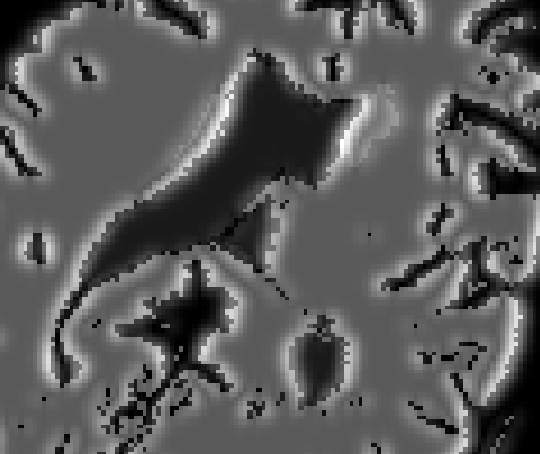
ANIMAL then estimates the non-linear spatial transformation required to match a pseudo US image volume to real US images acquired during surgery. This same transformation can be used to update the patient's MRI during surgery, thus permitting the neurosurgeon to make use of the pre-operative images during the intervention, even in the presence of a brain deformation (and errors in the linear registration from the patient to the pre-operative images).
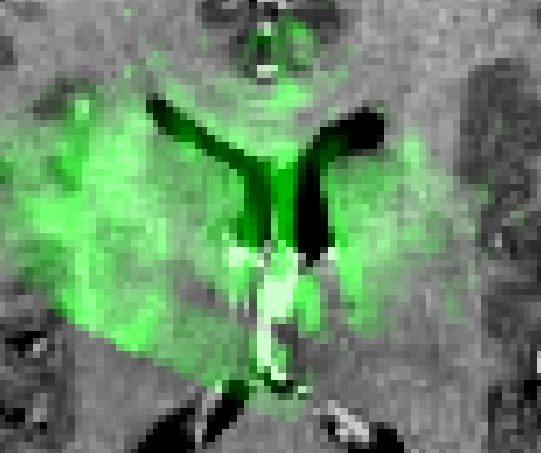
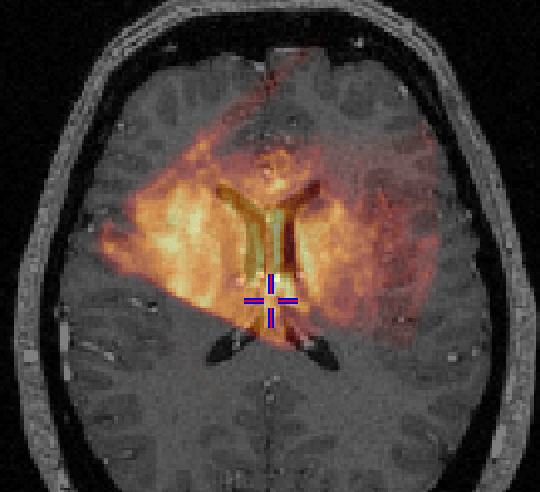
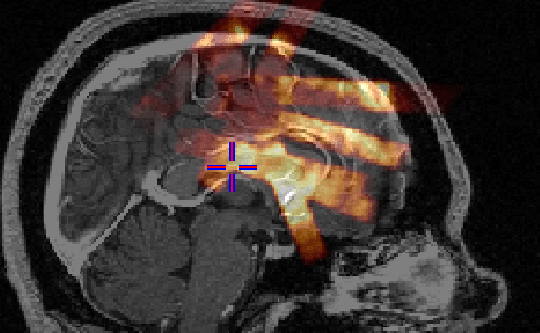
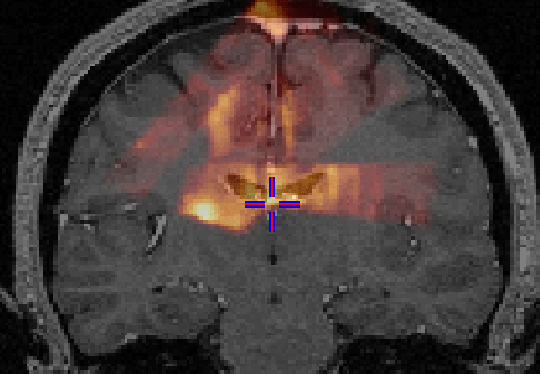
The feasibility of the approach is demonstrated through examination of the
qualitative results from two clinical cases. Figure 3 shows
the case of an amygdalo-hippocampectomy for intractable epilepsy. US images
were acquired at two different stages of the operation: before and after the
opening of the dura. The figure illustrates how the system is able to correct
for brain deformations at these two stages of surgery. The extent of the
correction can be seen in Figure 4. Figure 5 illustrates the case of a tumor
resection. Here, US images were acquired after the dura opening, when
significant brain deformation (on the order of 8mm) had occurred. The
strategy was able to correct for the deformations of both pathological and
anatomical structures. In all cases, it took approximately 30 seconds of
processing time to provide a corrected MRI volume on average.
 |
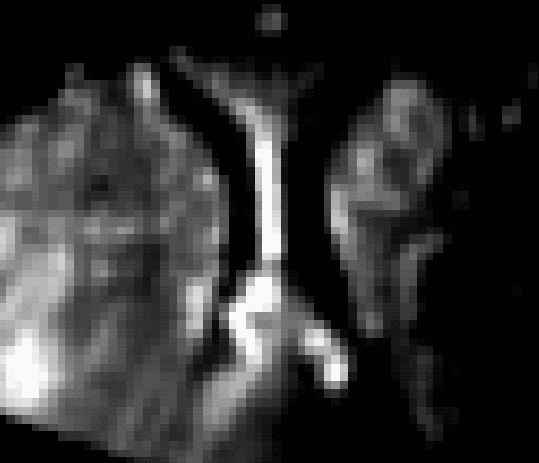 |
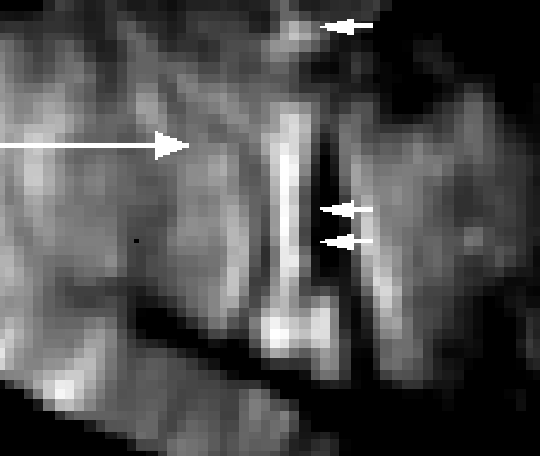 |
|---|---|---|
| Pseudo US | US before dura opening | US after dura opening |
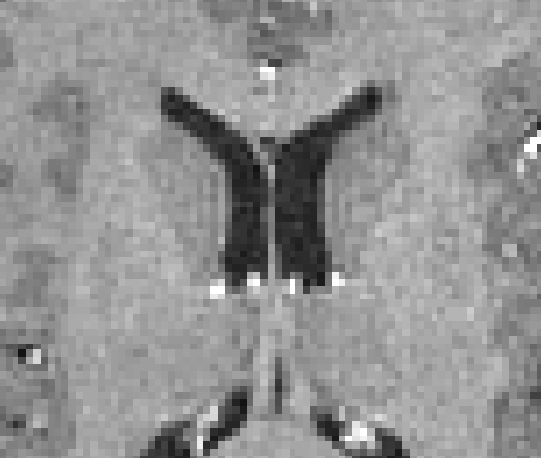 |
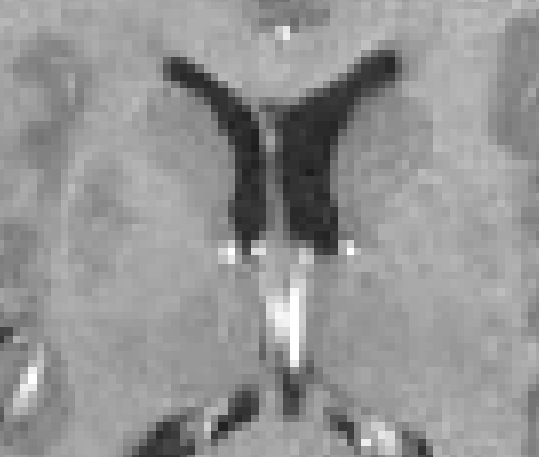 |
 |
| Original MRI | Corrected MRI | Corrected MRI |