BrainWeb:
Online Interface to a 3D MRI Simulated Brain Database
![[dag]](img3.gif)
Chris A. Cocosco, Vasken Kollokian, Remi K.-S. Kwan, Alan C. Evans
McConnell Brain Imaging Centre, Montréal Neurological Institute,
McGill University, Montréal, Canada
The increased importance of automated computer techniques for
anatomical brain mapping from MR images and quantitative brain image
analysis methods leads to an increased need for validation and
evaluation of the effect of image acquisition parameters on
performance of these procedures. Validation of analysis techniques of
in-vivo acquired images is complicated due to the lack of reference
data (``ground truth''). Also, optimal selection of the MR imaging
parameters is difficult due to the large parameter space. BrainWeb
makes available to the neuroimaging community, on-line on WWW, a set
of realistic simulated brain MR image volumes (Simulated Brain
Database, SBD) that allows the above issues to be examined in a
controlled, systematic way.
The SBD was generated by varying specific imaging parameters in an MRI
simulator, which starts from a digital phantom, and performs a
realistic, first-principles modeling of the imaging process based on
the Bloch equations [1]. The range of parameters was
chosen according to the values typically encountered in modern MRI
systems [2]. As an example of the generality of this
approach, MS lesions (extracted from real MRI-s) were added to the
normal brain phantom and the generation process was repeated. For
each anatomical model (phantom), three imaging sequences are available
online (T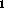 , T
, T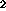 , PD), each with a fixed set of parameters:
typical values of slice thickness, noise and intensity non-uniformity
(INU) levels. All 3D image volumes are in stereotaxic space, and can
be interactively explored online in 3 simultaneous orthogonal views.
In addition, BrainWeb allows a remote user to run their own MRI
simulation through the WWW interface. Each simulated brain image, as
well as the source digital phantoms, can be downloaded in a variety of
file and data compression formats.
SBD can be used to study the performance of anatomical brain mapping
techniques, such as: non-linear co-registration [3],
cortical surface extraction, or tissue classification
algorithms [2].
Also, it can help the validation of quantitative analyses of
neuropathology (e.g. MS lesion quantification), or of other medical
pattern recognition and image processing techniques. The main
advantages of using this database are: (i) the answer is known a
priori in the experiment, and (ii) imaging parameters can be
independently controlled (see Fig. 1).
Since the source for all simulations is the same digital
phantom, one has a systematic means of establishing the sensitivity of
any particular procedure with respect to any imaging parameter or
artifact.
, PD), each with a fixed set of parameters:
typical values of slice thickness, noise and intensity non-uniformity
(INU) levels. All 3D image volumes are in stereotaxic space, and can
be interactively explored online in 3 simultaneous orthogonal views.
In addition, BrainWeb allows a remote user to run their own MRI
simulation through the WWW interface. Each simulated brain image, as
well as the source digital phantoms, can be downloaded in a variety of
file and data compression formats.
SBD can be used to study the performance of anatomical brain mapping
techniques, such as: non-linear co-registration [3],
cortical surface extraction, or tissue classification
algorithms [2].
Also, it can help the validation of quantitative analyses of
neuropathology (e.g. MS lesion quantification), or of other medical
pattern recognition and image processing techniques. The main
advantages of using this database are: (i) the answer is known a
priori in the experiment, and (ii) imaging parameters can be
independently controlled (see Fig. 1).
Since the source for all simulations is the same digital
phantom, one has a systematic means of establishing the sensitivity of
any particular procedure with respect to any imaging parameter or
artifact.
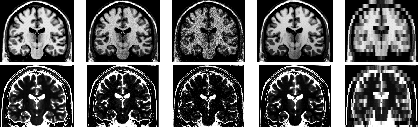
Figure: T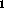 (top) & T
(top) & T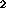 (bottom): (left to right)
ideal, typical, large noise, large INU, very thick slice
(bottom): (left to right)
ideal, typical, large noise, large INU, very thick slice
BrainWeb, containing the full SBD as well as the anatomical model
(phantom) used as input to the MR simulator, are available on WWW at
``http://www.bic.mni.mcgill.ca/brainweb/''.
We are currently working on extending SBD to include fMRI and PET simulated data.
References
- 1
-
Kwan, R.K.-S., Evans, A.C., Pike, G.B. In VBC,
Proceedings of the SPIE, 1996.
- 2
-
Kollokian, V.
Master's thesis, Concordia University,
Montreal, QC, Canada, Nov. 1996.
- 3
-
Collins, D.L., Holmes, C.J., Peters, T.M., Evans, A.C.
Human Brain Mapping. 3(3):190--208, 1996.
 Supported by the U.S. Human Brain Map Project and the International Consortium for Brain Mapping.
Supported by the U.S. Human Brain Map Project and the International Consortium for Brain Mapping.
Chris Cocosco (crisco@bic.mni.mcgill.ca)
May 8 1997
![[dag]](img3.gif)
![[dag]](img3.gif)
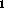 , T
, T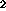 , PD), each with a fixed set of parameters:
typical values of slice thickness, noise and intensity non-uniformity
(INU) levels. All 3D image volumes are in stereotaxic space, and can
be interactively explored online in 3 simultaneous orthogonal views.
In addition, BrainWeb allows a remote user to run their own MRI
simulation through the WWW interface. Each simulated brain image, as
well as the source digital phantoms, can be downloaded in a variety of
file and data compression formats.
, PD), each with a fixed set of parameters:
typical values of slice thickness, noise and intensity non-uniformity
(INU) levels. All 3D image volumes are in stereotaxic space, and can
be interactively explored online in 3 simultaneous orthogonal views.
In addition, BrainWeb allows a remote user to run their own MRI
simulation through the WWW interface. Each simulated brain image, as
well as the source digital phantoms, can be downloaded in a variety of
file and data compression formats.

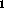 (top) & T
(top) & T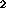 (bottom): (left to right)
ideal, typical, large noise, large INU, very thick slice
(bottom): (left to right)
ideal, typical, large noise, large INU, very thick slice Supported by the U.S. Human Brain Map Project and the International Consortium for Brain Mapping.
Supported by the U.S. Human Brain Map Project and the International Consortium for Brain Mapping.