



Next: 3D Results - Neck
Up: Results
Previous: Synthetic Labelled Images
The next exercise attempted to reduce deformation at an area where source-target differences are known a priori in order to highlight other areas where there are unknown differences due to abnormalities. The target and source were coronal slices of a pre-/post-operative data set exhibiting hydracephalous and coning, Fig. 4. Since these are slices through the same subject at approximately, but not exactly, the same location, they exhibit slight differences in scalp shape. To some extent the same applies to the cortex and internal brain structures; however the main cause of their source-target differences was the surgical procedure and its after-effects. It is these differences which were to be highlighted as abnormal.
Figure 4:
Data set of ( left) head4A and ( right) head4B
![\begin{figure}\makebox[\textwidth]{
\epsfxsize=0.2\textwidth \epsfbox{hen65pre.ps}
\epsfxsize=0.2\textwidth \epsfbox{hen65post.ps}
}
\end{figure}](img37.gif) |
We compared registration by the UF, by MF3
and by MF2.
The target (pre-operative) image was used as an atlas, defining the normal brain shape for that subject. The scalp region was segmented manually with the aid of the display tool xdispunc developed by Dave Plummer of UCH Medical Physics. This prior information of known `abnormal' scalp shape was supplied as a binary image indicating the region where deformation was to be reduced. Five sets of images were generated for each registration:
- 1.
- the deformed source image,
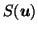 after registration to the target T.
after registration to the target T.
- 2.
- the same deformation applied to a regular grid image,
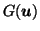 .
The ideal inhomogeneous registration paradigm would exhibit no deformation in
.
The ideal inhomogeneous registration paradigm would exhibit no deformation in
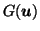 in the scalp region (painted white on the grid prior to deformation).
in the scalp region (painted white on the grid prior to deformation).
- 3.
- local magnitudes of the resulting displacements field and local Laplacian, bending and elasticity energy metrics [12], to highlight regions of severe distortion of the source image from the normal brain shape.
- 4.
- the difference of the deformed source from the undeformed source,
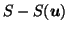 .
Assuming registration to the target is optimal in regions where no prior information was supplied, this gives a further indication of abnormality, defined as shape difference from the target. (Known) differences in the scalp region are not highlighted if its registration to the target has been successfully suppressed.
.
Assuming registration to the target is optimal in regions where no prior information was supplied, this gives a further indication of abnormality, defined as shape difference from the target. (Known) differences in the scalp region are not highlighted if its registration to the target has been successfully suppressed.
- 5.
- the difference of the deformed source from the target,
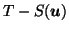 .
This is to check that registration is complete in the unknown regions and suppressed in the scalp. In this case, for an ideal registration, the difference image is zero in unknown regions and non-zero in the scalp region.
.
This is to check that registration is complete in the unknown regions and suppressed in the scalp. In this case, for an ideal registration, the difference image is zero in unknown regions and non-zero in the scalp region.
The results are shown in Fig. 5. Of the three registrations, in
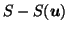 ,
MF2 with motionless scalp is the clearest at highlighting only the differences in the ventricular and left-cortical areas, (Fig. 5, bottom row, far right). By inspecting the grid lines in the white regions of
,
MF2 with motionless scalp is the clearest at highlighting only the differences in the ventricular and left-cortical areas, (Fig. 5, bottom row, far right). By inspecting the grid lines in the white regions of
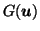 (Fig. 5, bottom row centre left) we see it has respected the rigidity of the scalp. Finally, to check completeness of registration, Fig. 5, (bottom row centre right) shows good registration in the brain region (the difference image shows little structure) and poor registration at the scalp.
In comparison, MF3 allowed the scalp to distort, shown in the grid image (Fig. 5, centre row, centre left), leaving less structure in the scalp region of
(Fig. 5, bottom row centre left) we see it has respected the rigidity of the scalp. Finally, to check completeness of registration, Fig. 5, (bottom row centre right) shows good registration in the brain region (the difference image shows little structure) and poor registration at the scalp.
In comparison, MF3 allowed the scalp to distort, shown in the grid image (Fig. 5, centre row, centre left), leaving less structure in the scalp region of
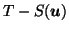 (Fig. 5, centre row, centre right); hence the known scalp shape difference shows up as an abnormality in
(Fig. 5, centre row, centre right); hence the known scalp shape difference shows up as an abnormality in
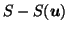 (Fig. 5, centre row, far right).
(Fig. 5, centre row, far right).
Figure:
Results of registering source S = head4B to target T = head4A. ( Rows, top-bottom): UF;
MF3;
MF2. ( Columns, left-right): deformed S, S(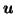 ); deformation applied to grid, G(
); deformation applied to grid, G(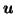 ); T - S(
); T - S(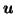 ), showing completeness of registration; S - S(
), showing completeness of registration; S - S(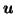 ), highlighting differences due to the deformation ( right 2 columns are contrast-enhanced)
), highlighting differences due to the deformation ( right 2 columns are contrast-enhanced)
![\begin{figure}\makebox[\textwidth]{
\epsfxsize=0.2\textwidth \epsfbox{Lou_rT.ps}...
...fbox{Lou_popr.ps}
\epsfxsize=0.2\textwidth \epsfbox{Lou_popo.ps}
}
\end{figure}](img43.gif) |
Figure 6:
Deformation metrics ( left-right): Laplacian, bending, elastic, magnitude of transformation, of the registration by ( top) UF and ( bottom) MF3
![\begin{figure}\makebox[\textwidth]{
\epsfysize=0.2\textwidth \epsfbox{Lou_rL.ps}...
...\epsfbox{Lou_ve.ps}
\epsfysize=0.2\textwidth \epsfbox{Lou_vt.ps}
}
\end{figure}](img44.gif) |




Next: 3D Results - Neck
Up: Results
Previous: Synthetic Labelled Images
Hava LESTER
1999-03-24
![]() ,
MF2 with motionless scalp is the clearest at highlighting only the differences in the ventricular and left-cortical areas, (Fig. 5, bottom row, far right). By inspecting the grid lines in the white regions of
,
MF2 with motionless scalp is the clearest at highlighting only the differences in the ventricular and left-cortical areas, (Fig. 5, bottom row, far right). By inspecting the grid lines in the white regions of
![]() (Fig. 5, bottom row centre left) we see it has respected the rigidity of the scalp. Finally, to check completeness of registration, Fig. 5, (bottom row centre right) shows good registration in the brain region (the difference image shows little structure) and poor registration at the scalp.
In comparison, MF3 allowed the scalp to distort, shown in the grid image (Fig. 5, centre row, centre left), leaving less structure in the scalp region of
(Fig. 5, bottom row centre left) we see it has respected the rigidity of the scalp. Finally, to check completeness of registration, Fig. 5, (bottom row centre right) shows good registration in the brain region (the difference image shows little structure) and poor registration at the scalp.
In comparison, MF3 allowed the scalp to distort, shown in the grid image (Fig. 5, centre row, centre left), leaving less structure in the scalp region of
![]() (Fig. 5, centre row, centre right); hence the known scalp shape difference shows up as an abnormality in
(Fig. 5, centre row, centre right); hence the known scalp shape difference shows up as an abnormality in
![]() (Fig. 5, centre row, far right).
(Fig. 5, centre row, far right).