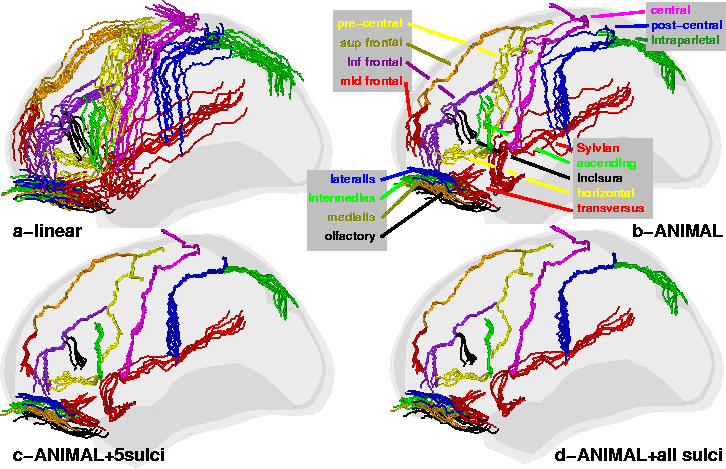
Standard ANIMAL: In the first experiment (Fig.
2-b) we can see that the global brain shape is properly
corrected and that the main lobes are mostly aligned since the central and
Sylvian sulci are better aligned than in the linear registration. Even
though previous experiments in [2] have shown that basal
ganglia structures (e.g., thalamus, caudate, putamen, globus pallidus) and
ventricular structures are well registered (with overlaps on the order of
85% to 90%) for both simulated and real data, the simulations presented
here indicate that the standard ![]() technique cannot always register
cortical structures by using only blurred image intensities and gradient
magnitude features.
technique cannot always register
cortical structures by using only blurred image intensities and gradient
magnitude features.
ANIMAL+selected sulci: In Fig. 2-c, 10
extracted sulci (superior and middle frontal, Sylvian, olfactory and
central, on both hemispheres) were used as additional features in ![]() to address problems in establishing correspondence. Visual inspection of
Fig. 2-c shows that the previous misalignments in the
motor-sensory area have been corrected, and alignment for neighbouring
structures has improved greatly. Indeed, when evaluated on the central
sulcus, the
to address problems in establishing correspondence. Visual inspection of
Fig. 2-c shows that the previous misalignments in the
motor-sensory area have been corrected, and alignment for neighbouring
structures has improved greatly. Indeed, when evaluated on the central
sulcus, the
![]() measure improves dramatically: 3.2mm for linear;
2.0mm for standard non-linear; 0.7mm for
measure improves dramatically: 3.2mm for linear;
2.0mm for standard non-linear; 0.7mm for ![]() with labelled sulci.
The standard deviation of
with labelled sulci.
The standard deviation of
![]() decreases as well, indicating tighter
grouping around the target sulci. The 0.54mm average
decreases as well, indicating tighter
grouping around the target sulci. The 0.54mm average
![]() reduction is
highly significant (p<0.0001, T=157,
d.o.f.=320, paired T-test over all sulci and all
subjects).
reduction is
highly significant (p<0.0001, T=157,
d.o.f.=320, paired T-test over all sulci and all
subjects).
ANIMAL+selected sulci+all sulci: In this experiment, the
![]() +selected sulci transformation is used as input where all sulci
extracted by
+selected sulci transformation is used as input where all sulci
extracted by ![]() are used as features in an attempt to further improve
cortical alignment in regions that are not close to the previously selected
sulci. The improvement is evident qualitatively in Fig. 2-d
and quantitatively in Table 1 where the average
are used as features in an attempt to further improve
cortical alignment in regions that are not close to the previously selected
sulci. The improvement is evident qualitatively in Fig. 2-d
and quantitatively in Table 1 where the average
![]() ,
evaluated over all
sulci and all subjects drops from 1.6mm for the standard
,
evaluated over all
sulci and all subjects drops from 1.6mm for the standard ![]() to
1.5mm when using all sulci. While this improvement is small, it is
statistically significant (p<0.0001, T=88.5,
d.o.f.=320, paired T-test).
to
1.5mm when using all sulci. While this improvement is small, it is
statistically significant (p<0.0001, T=88.5,
d.o.f.=320, paired T-test).