Ovine Brain Atlas
Overview
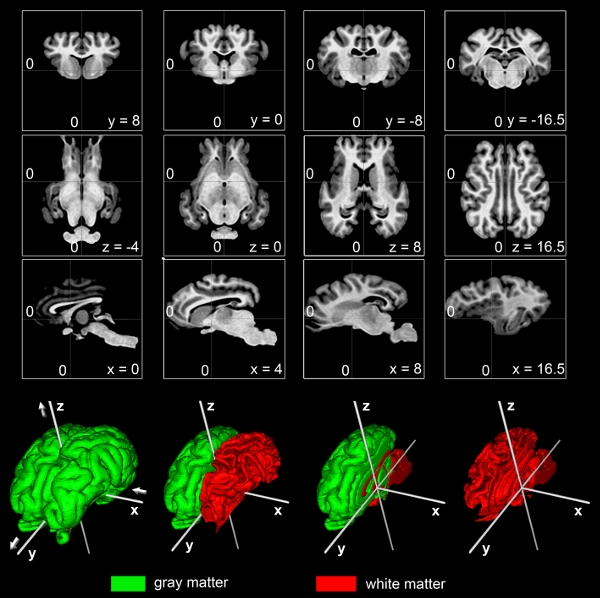
We present an ovine, unbiased, population-averaged standard magnetic resonance imaging template brain volume that offers a common stereotaxic reference frame to localize anatomical and functional information in an organized and reliable way for comparison across individual sheep and studies. We have used t1w MRI volumes from a group of 14 normal adult sheep to create the template and a priori probability of cerebral gray (GM) and white (WM) matter as well as cerebrospinalfluid (CSF). Thus, the atlas does not rely on the anatomy of a single subject, but instead depends on nonlinear normalization of numerous sheep brains mapped to an average template image that is faithful to the location of anatomical structures. Tools for registering a native MRI to the ovine space can be found in the Software section.
Methods
The ovine average atlas is comprised of 14 T1-weighted MRIs of normal young adult sheep brains (Ovis orientalis aries). This atlas is not based on a single subject but instead is an average constructed from the averaged position, orientation and scale from all the individual subjects and is representative of both the intensities and spatial positioning of anatomical structures. Image pre-processing included non-uniform intensity correction and intensity normalization with a range of 0ñ100. One out of 14 sheep was chosen to serve as the initial target. Registration of the remaining 13 sheep brains to the chosen was initialized using manually identified homologous landmarks that included the center of the left/right eyeball, the most rostral and caudal poles of both hemispheres, the intersection of the cruciate sulcus and the longitudinal fissure, as well as the anterior (AC) and posterior commissure (PC). A three-step average template generation process (Fonov et al., 2009) was used to estimate the nonlinear deformation field that best aligned local neighbourhoods between each sheep brain volume and the average target. In brief, the transformation matrices calculated in one step were applied to the appropriate scan in order to resample the image and where then used for the next step. The following processing steps were performed: a linear left-right symmetric rigid body registration using 8 iterations. The rigidly transformed average was created. Then, a left-right symmetric fully-affine (12 parameters) iterative registration procedure was applied to each input scan using 8 iterations followed by calculation of the average. A left-right symmetric non-linear registration including 4 iterations with 4 mm steps, 4 iterations with 2 mm steps and 4 iterations with 1 mm steps was applied to each scan before the final, non-linear transformed average was generated.
Publications
The following publications should be referenced when using this atlas:
Nitzsche B, Frey S, Collins LD, Seeger J, Lobsien D, Dreyer A, Kirsten H, Stoffel MH, Fonov VS and Boltze J (2015). A stereotaxic, population-averaged T1w ovine brain atlas including cerebral morphology and tissue volumes. Front. Neuroanat. 9:69. doi: 10.3389/fnana.2015.00069
Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC& Collins DL (2011). Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 54 (1), 313–327.
Download
Click on the appropriate resolution and file format below to download archive:
0.5mm resolution: MINC1 27Mb, MINC2 26Mb, NIFTI 48Mb
0.25mm resolution: MINC1 204Mb, MINC2 197Mb, NIFTI 413Mb
Sample dataset in NIFTI format: 24Mb
Brain surfaces (STL format): 20Mb
Contact
For questions related to the MNI macaque atlas (rather than the website), contact Björn Nitzsche bjoern.nitzsche <at> izi.fraunhofer.de
