Overview
CIVET is a human brain image-processing pipeline for fully-automated corticometric, morphometric and volumetric analyses of magnetic resonance (MR) images. The pipeline is composed of a set of Perl scripts and high-level C/C++ programs.
CIVET starts by a 9 or 12-parameter linear/affine registration of the MR images (T1-weighted, T2-weighted and Proton Density weighted) from native to stereotaxic space, using the average MNI ICBM152 model (or any other volumetric model that is available) as the target of registration. Once the images have been transformed to stereotaxic space, the images are corrected for radio-frequency non-uniformities and a brain mask is computed. A non-linear transformation is then computed from the subject in stereotaxic space to the model.
The next step in CIVET is to perform the tissue classification into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF), either based on the T1-weighted image only, or on multispectral data (T1, T2, PD). The classification is comprised of two steps: a discrete classification based on tag points, followed by the evaluation of the partial volumes for the tissue classes.
The brain is split into the left and right hemispheres for the purpose of surface extraction. The boundary between cortical GM and subcortical WM (referred to as the white matter surface) is first extracted. The pial surface, or the boundary betwen the cortical GM and the extra-cortical CSF (referred to as the GM surface) expands outwards from the WM surface to the CSF. Each default surface (per hemisphere) is composed of 81,920 triangles (polygons) and 40,962 vertices. High resolution surfaces are also available at 327,680 polygons and 163,842 vertices.
The surfaces are registered to the MNI ICBM152 surface template (or any other surface model that is available). Surface registration as such allows for group comparisons by interpolating vertex-based cortical measurements at reference vertices on the template. The cortical thickness is computed by evaluating the distance, in mm, between the original WM and GM surfaces transformed back to the native space of the original MR images, then interpolated onto the surface template. Vertex-based areas and volumes are, on the other hand, computed directly on the resampled surfaces and measure local variations of area/volume contraction and expansion relative to the vertex distribution on the surface template.
Regional maps are produced based on a lobar parcellation of the surfaces (major lobes, AAL, or DKT-40). Other measurements such as mean curvature, gyrification index, and total cortical area are also computed.
A number of figures, tables, and reports are automatically produced to assist in the process of quality control and data analysis.
What’s new in CIVET-2.0.0?
The major improvements are outlined below.
- Marching-cubes algorithm:
- White surface t1-gradient calibration:
- New N3 version:
- High-resolution surfaces:
- Surface blurring fwhm:
- Mean curvature:
- Classified tissue volumes:
- ANIMAL segmentation:
Some consequences of these major improvements are:
- Reduced dependence of results on the N3 spline distance:
- New average population surfaces:
Overall, CIVET-2.0.0 provides a major overhaul of the surface extraction and associated algorithms to provide high-quality cortical surfaces and more precise measurements of cortical thickness. New functionalities have been added, for instance: processing at different voxel sizes, high-resolution surfaces, ANIMAL segmentation, improved quality control tools, etc. The user should also find this new version easier to use since results are mostly invariant to the N3 spline distance. Finally, fewer subjects are lost due to processing failures (e.g. mis-registration).
Finally, the computational resources required by CIVET 2.0.0 are shown below. At comparable conditions (1mm voxel size and low-res surfaces), CIVET 2.0.0 offers a reduction in disk space due to the elimination of some unnecessary intermediate files and cropping of the non-linear transformation deformation field. In terms of computational time, CIVET 2.0.0 is very competitive throughout the various options, from low-res to hi-res surfaces at different voxel size resolution. (In the figures, the suffix _mc means marching-cubes for CIVET 2.0.0.)
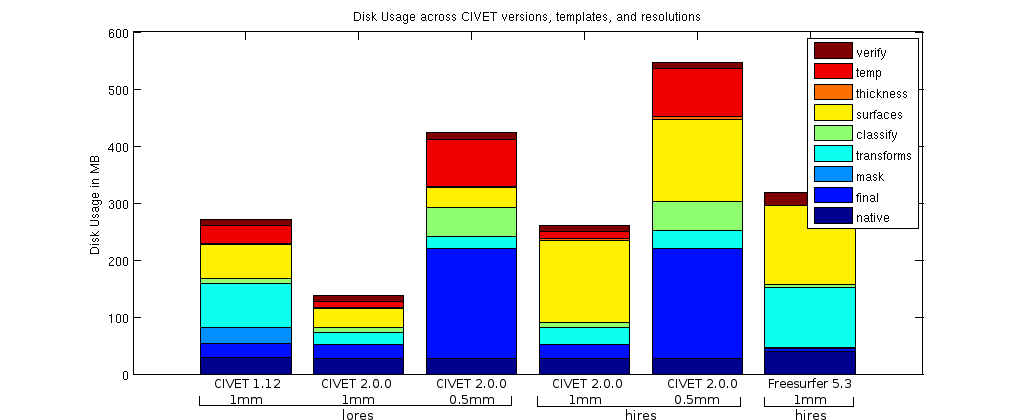
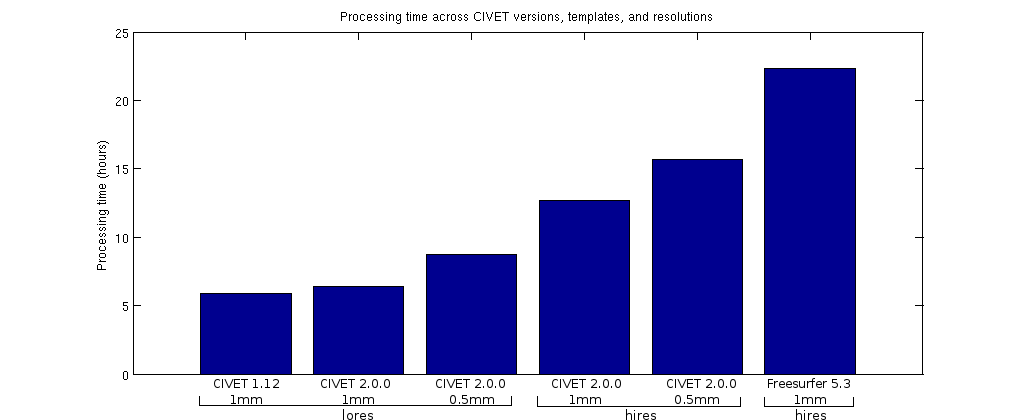
Full summary of major changes between CIVET 1.1.12 and CIVET 2.0.0:
*************************************************************************
New in September, 2014 quarantine, version 2.0.0
1 - marching-cubes algorithm for extraction of initial white surface
2 - new calibration of white surface at GM-WM t1-gradient
3 - improved node movement in surface fit program
4 - corrected functional for Laplacian constraint in surface
fit program (for gray surface)
5 - improved surface registration
6 - new surface registration models based on marching-cubes
7 - simplified CIVET QC pipeline
8 - corrected pve classification at 0.50mm volume template
9 - all changes listed below for 1.1.13
10 - ANIMAL volumetric lobar segmentation
11 - corrected fwhm for surface smoothing (was off by sqrt(2) in 1.1.12)
Note: This version requires the package surface-extraction-3.0.2
or higher.
More to come soon:
1 - classification and surface extraction in native space.
2 - intermediate model for linear and non-linear volume registration.
3 - Plug-in for alternate population templates
4 - removal of self-intersections in .sm file for surface
registration (Maxime Boucher)
5 - Longitudinal surface registration and QC
*************************************************************************
New in July, 2013 quarantine, version 1.1.13 (not released)
1 - bug fixed in N3 to be invariant to voxel size (requires N3-1.12.0
and EBTKS-1.6.4)
2 - high-resolution surfaces at 320k polygons
3 - surface registration on high-resolution surfaces, true
multi-resolution algorithm (faster)
4 - improved medial cut through the corpus callosum
5 - processing at voxel sizes of 0.5mm and 1.0mm
6 - speed optimizations for pve
7 - speed optimizations for Laplacian field at 0.5mm
8 - cropping of non-linear xfm grid file to save disk space
and to reduce peak memory
9 - masking of hippocampus and amygdala for icbm152nl_09s model (0.5mm)
10 - masking of hippocampus and amygdala for ADNIhires model (0.5mm)
11 - improved schedule for white matter surface extraction
(smoother convergence, scaled starting ellipsoid)
12 - new ICBM surface average for scaled ellipsoid
13 - clean-up of connected pieces of white matter (partial
volumes)
14 - new pipeline to build average surfaces from CIVET output
