This page is under construction.
Introduction
CIVET-macaque is based on CIVET-2.1.1 for the human, adapted to the macaque. CIVET-chimp, as the name says, is adapted to the chimpanzee. New users should read the CIVET documentation to get familiar with the CIVET terminology, features, inputs, outputs, etc.
Running CIVET-macaque and CIVET-chimp on CBRAIN
- CIVET-macaque/chimp (version alpha-0.2) is current accessible via CBRAIN.
- Configuration
- spline distance: this depends on scanner field strength (range 50 to 125)
- head height: distance from the top of the scalp to the bottom of the cerebellum. This distance needs to be larger for male subjects due to thicker scalp. Default is 50mm (macaque) and 110mm (chimpanzee), but it might be safer to set explicitly to 0.
- recommended voxel size: should be 0.25mm for the macaque and 0.40mm for the chimpanzee.
- NIfTI: CIVET-macaque can automatically read inputs in minc or NIfTI formats. Optionally, you can save a copy of the results in NIfTI format, for your convenience. The surfaces and surface maps will be saved in GIfTI format, with full compatibility with HCP Work Bench tool wb_view.
- parcellations: only one surface parcellation can be chosen in a given run (macaque: lobes, D99, CHARM; chimp: lobes, Davi130, Brainnetome).
Common errors
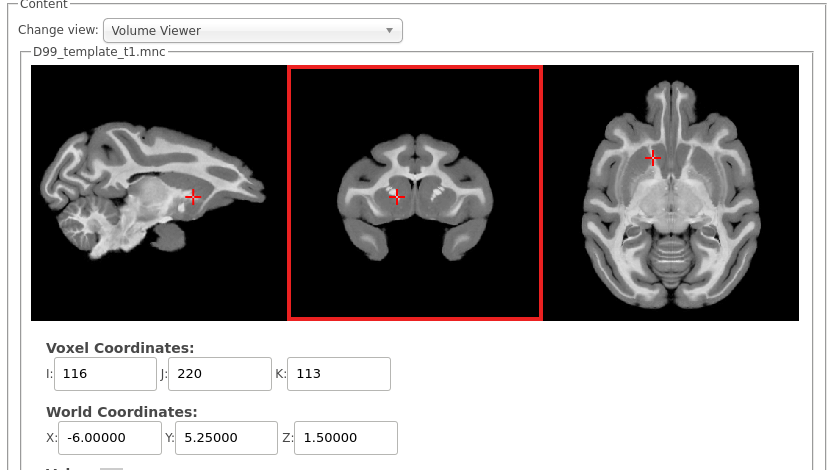
- Scan orientation: the scan must be acquired in the sphynx position. When viewing the volume in the volume viewer in CBRAIN, the volume must look like below. 1) the eyes must point upward on the axial view (3rd column); and 2) the eyes must point to the right on the sagittal view (first column). On the coronal view (middle column), left is left and right is right (neurological view). An incorrect scan orientation will always lead to a failure in CIVET. The volume must thus be rotated in such a case before processing. Note that the input volume does not need to be pre-masked.
- Masking: if the scans are pre-masked, select that option in the CBRAIN configuration
- Scan quality: CIVET-macaque/chimp is quite sensitive to scan quality — contrast, SNR, bias field, voxel size, visible blood vessels. Don’t expect miracles.
- Registration: male/female head size differences (scalp) can mislead registration. The head height parameter may need to be set differently for males vs females.
- Surface extraction: This is the most common error in CIVET — incorrect topology of the surface after surface-based registration. There is no need to re-run multiple times. If there are self-intersections, re-running will not help. Look for visible blood vessels, mis-registration, scan quality, non-uniformity corrections, etc. As a general rule, the outputs will look no better than the inputs.
Working with the minc tools
- You can download the minc toolkit to obtain the minc tools.
- Transforming a CIVET surface from stereotaxic space to native space:
- xfminvert transforms/linear/t1_tal.xfm t1_tal_inv.xfm
- transform_objects surfaces/white.obj t1_tal_inv.xfm surfaces/white_native.obj
- Intersecting a surface with a volume in native space:
- volume_object_evaluate -linear native_volume.mnc native_surface.obj volume_field.txt
- Viewing volumes:
- register volume1.mnc
- register volume1.mnc volume2.mnc
- Display volume1.mnc
- Viewing surfaces on volumes:
- Display volume1.mnc white_left.obj gray_left.obj
- Viewing surfaces with BrainBrowser
- BrainBrowser is web-based. Load a surface and its surface field. Many formats are available.
Surface-based registration, average surface and atlases
The surfaces are extracted in the species-specific stereotaxic space and registered to the average surface for the NMT or NCBR dataset for the macaque or chimpanzee, respectively. Surface-based registration is necessary for group-based and ROI-based analyses by sampling the surface (placement of the vertices) at the same morphological positions across subjects. Resampled surface-based registered surfaces (and surface maps files) have the suffix _rsl_ in their names. These are the files to use for group analyses and to view labels from a parcellation atlas.
You can download the CIVET-macaque average surface (version alpha-0.1) or CIVET-macaque average surface (version alpha-0.2) on which to display your group data, along with the D99 and CHARM parcellations for group-based and ROI-based analyses.
You can download the CIVET-chimp associated files archive (version alpha-0.2), which contains the NCBR volumetric template and the average population surface, along with the Davi130 and Brainnetome parcellations for group-based and ROI-based analyses.
The Matlab SurfStat package can be used for statistical analyses of CIVET results.
References for CIVET-macaque
You must cite the main CIVET-macaque paper.
NeuroImage 227, 2021.
together with the paper describing the NIMH Macaque Template (NMT v1.2) used in CIVET-macaque
If you are using the D99 parcellation atlas, you must additionally cite
If you are using the CHARM parcellation atlas, you must addtionally cite
References for CIVET-chimp
You must cite the main CIVET-chimp paper (submitted to NeuroImage).
If you are using the Davi130 parcellation atlas, you must additionally cite
If you are using the Brainnetome parcellation atlas, you must addtionally cite
References for CBRAIN
Finally, if you are using CBRAIN, you must also cite CBRAIN and acknowledge Digital Research Alliance of Canada (formerly Compute Canada) for the usage of the computing resources.
