Once all subjects have processed to completion, it is necessary to perform quality control (QC).
If you used Cbrain to process your data, you should have already run the QC pipeline by checking the box at the end for “Launch Claude’s QC script.” If you are using the command line, you may run the CIVET QC pipeline as follows:
CIVET_QC_Pipeline -sourcedir <dir> -targetdir <dir> -prefix <prefix> <id1> <id2> … <idn> <idfile> > <logfile>
CIVET_QC_Pipeline -sourcedir <dir> -targetdir <dir> -prefix <prefix> -id-file <idfile> > <logfile>
The output of this pipeline is located in the target directory targetdir/QC. Important files to look for are:
civet_$prefix.html: main HTML file to load in your web browsercivet_$prefix.glm: generic GLIM file corresponding to the main HTML that can be viewed as a spreadsheet (*.csv) and/or imported into SurfStat for analysis
In the HTML table, orange boxes indicate a warning while red boxes indicate possible errors. The results of red boxed subjects should be examined carefully.
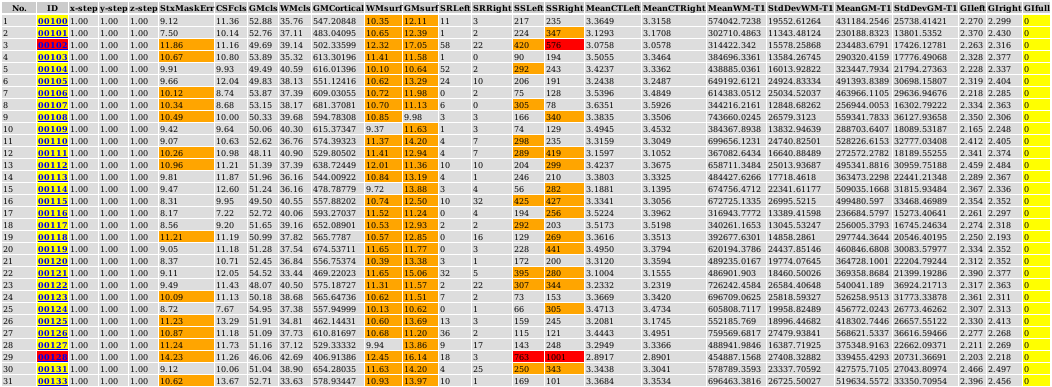

Various metrics are tabulated for the subjects:
x-step, y-step, z-step: voxel size of the native t1 imageStxMaskErr: error in brain mask in stereotaxic space (difference between subject’s mask and model’s mask), should be 10% or lessCSFcls, GMcls, WMcls: percentages of classified CSF, GM, WM in masked imageGMCortical: volume of cortical gray matter in mm^3 (excludes sub-cortical gray)WMsurf: percentage error in extraction of white surface (should be 10% or less), defined as the number of classified WM voxels outside the white surface (inside the brain mask) divided by the number of voxels of the brain mask.GMsurf: percentage error in expansion of gray surface (should be 15% of less), defined as the number of GM voxels outside the gray surface (inside the brain mask) divided by the number of voxels of the brain mask.SRLeft, SRRight: number of self-intersections of left and right resampled mid-surfaces (should be 100 or less)SSLeft, SSRight: number of surface-surface intersections between left and right white and gray surfaces (values above 500 are often associated with defects due to motion artifacts or masking)MeanCTLeft, MeanCTRight: mean cortical thickness for left and right hemispheres (values much below the population average are often associated with defects due to motion artifacts)MeanWM-T1, StdDevWM-T1: mean and standard deviation of t1 intensities for pure class of classified white matterMeanGM-T1, StdDevGM-T1: mean and standard deviation of t1 intensities for pure class of classified gray matterGIleft, GIright, GIfull: gyrification index of gray surface for left, right and combined hemispheres
At the bottom of this table, the population mean and standard deviation is given for most of these fields. Outliers can be identified by values more than one standard deviation away from the mean, as seen in the plots following the table.
By clicking on the subject id in the table, you can view its verify .png images.
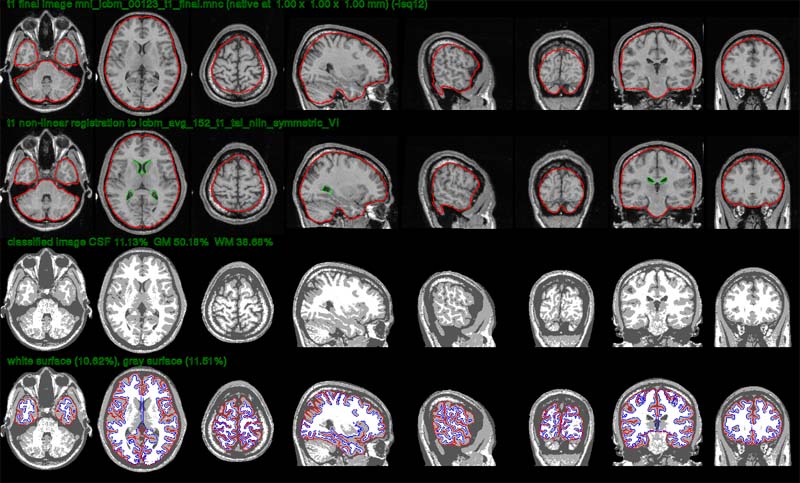
In the verify image below, one looks at the quality of the registration. The first row shows the linearly registered subject, with the red outline representing the border of the brain mask for the stereotaxic model. The subject’s brain should fit tightly inside the red outline for a perfect linear registration. The second row shows the non-linearly transformed subject to the stereotaxic model. The green outline represents the ventricles of the model. The third row shows the tissue classification while the last row shows the white (blue) and gray (red) cortical surfaces overlaid on top of the tissue classification. The outline of the cortical surfaces should match the tissue classification (zoom in a bit) for complete surface extraction.
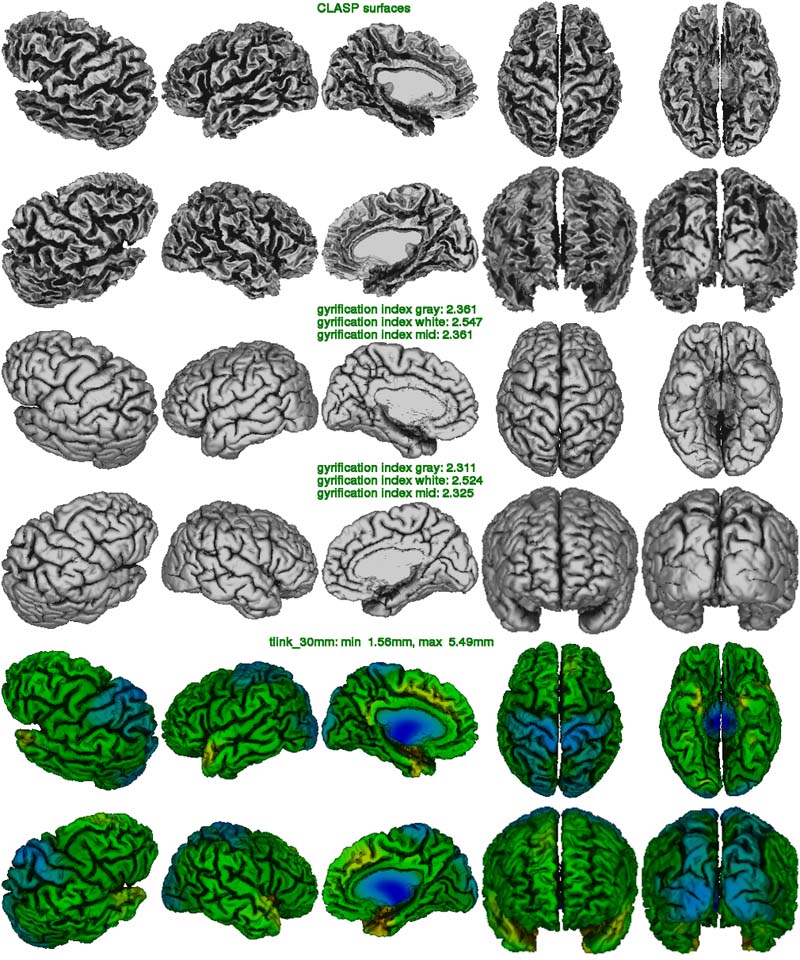
In the clasp image below, one looks at the quality of the surface extraction. (Note: due to maximum size restriction in this web documentation, the resolution of this figure has been reduced.) This figure is helpful to detect masking errors (pieces of skull attached to white surface), motion artifacts (white surface looks jaggy with many bridges across gyri), under-expansion of the gray surface (often in frontal and temporal lobes), as well as poor N3 correction of non-uniformities (atypical pattern of cortical thickness).
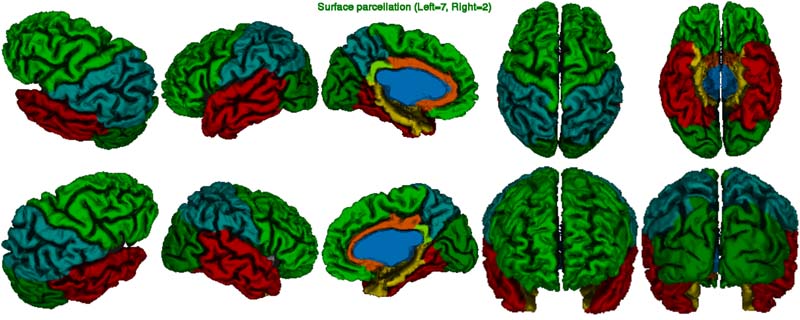
In the atlas image below, one looks at the quality of the surface registration. In particular, the border of the frontal and parietal lobes at the central sulcus must be perfectly identified. The AAL atlas is also available as an option in CIVET.
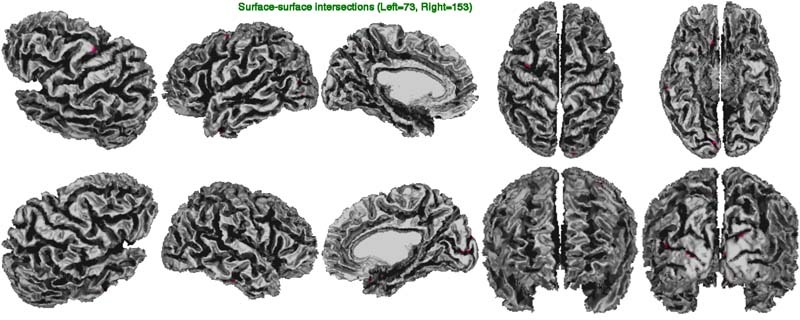
In the surfsurf image below, one looks at the number of gray-white surface-surface intersections (tiny spots in pink). These usually occur when the gray surface is arbitrarily close to the white surface as a result of abnormally thin cortex (usually indicative of poor N3 correction of non-uniformities), of masking errors, of tissue classification errors (often, broken thin strips of white matter in the temporal lobes), and of defects in white surface extraction (erroneous bridge across a sulcus cutting through cortex).
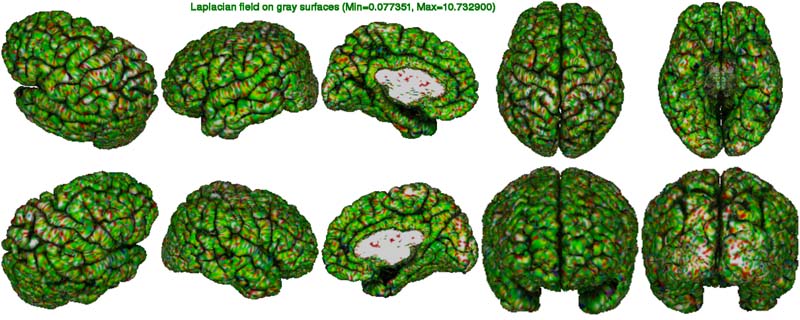
In the laplace figure below, one looks at the convergence of the expansion of the gray surface. A totally green image (Laplacian value of 10) indicates perfect convergence. Colours in high spectrum (reg to white) indicate vertices outside the
pial surface (over-expansion) while colours in the low spectrum (blue to black) indicate vertices inside the cortex (under-expansion). The merits of this figure will become apparent in a future version of CIVET based on marching-cubes.
Examples of QC failure
Brainbrowser
You may also load individual volumes or surfaces into Brainbrowser for additional inspection. Brainbrowser allows for visualization of volumes and surfaces through a standard web browser, without the need for specialty software installation.
https://brainbrowser.cbrain.mcgill.ca/
In the event that you suspect motion artifacts, you may wish to use the Volume Viewer in order to scroll through your volume (*.mnc file) to search for potential “ringing.”
https://brainbrowser.cbrain.mcgill.ca/braincanvas
Or, you may wish to use the Model Viewer in order to view your surface (*.obj file) from additional angles.
https://brainbrowser.cbrain.mcgill.ca/surfview.html
